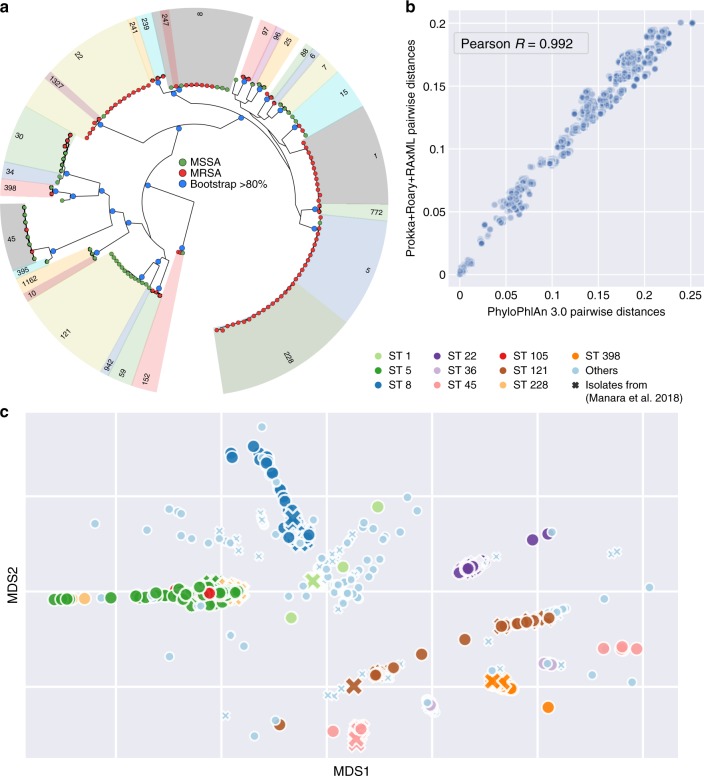
Fig. 2. Accurate reconstruction of Staphylococcus aureus phylogenies using PhyloPhlAn 3.0.
a Phylogenetic tree of 135 S. aureus strains from a pediatric hospital36 reconstructed by PhyloPhlAn 3.0 using 2127 automatically identified core genes (rendered by GraPhlAn39 see Supplementary Fig. 2 for a full comparison). Green circles represent the methicillin-sensitive S. aureus (MSSA), while red circles represent methicillin-resistant S. aureus (MRSA). Blue circles internal to the phylogeny identify subtrees with bootstrap >80%. b Normalized phylogenetic distances in the PhyloPhlAn 3.0-reconstructed tree and in a manually curated phylogeny from ref. 36 highlighting strong consistency between the automated PhyloPhlAn 3.0 results and the curated tree (0.992 Pearsonʼs correlation coefficient). c Multidimensional scaling ordination of pairwise phylogenetic distances from the tree integrating the 135 S. aureus isolates (crosses) with 1000 automatically selected S. aureus reference genomes (circles, Supplementary Fig. 1). The ten most prevalent sequence types (STs)23 are highlighted in different colors.