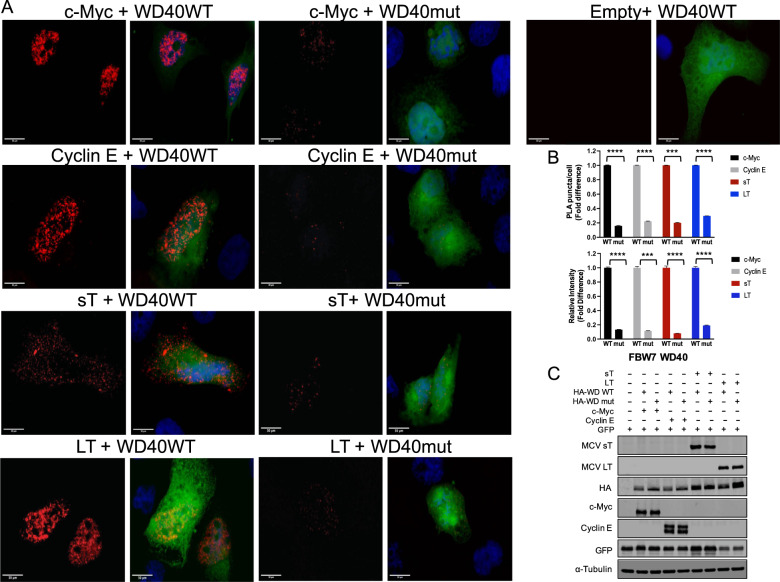
Fig. 3. A comparative analysis of FBW7 interaction by proximity ligation assay.
a MCV LT, c-Myc, Cyclin E and MCV sT were co-expressed with either FBW7ΔDF (WT) or V375A/K377N (mut) in U2OS cells. At 48 h post transfection, a proximity ligation assay (PLA) was performed. Technical controls (Empty vector and FBW7ΔDF (WT)) demonstrate the specificity of PLA signals in samples. Representative images for each sample were prepared by uniformly adjusting brightness and contrast for ImageJ analysis. Red dotted signal indicates interaction and displayed separately. Nuclei were stained with DAPI. GFP plasmid was co-transfected as a transfection control for all samples. GFP fluorescence also indicates the perimeter of the cell. (Scale bar = 30 μm). b Quantification of PLA signals. Protein interactions were quantified by counting the number of puncta per cell as well as measuring the florescence intensity of puncta signals per cell. The dots (indicating interactions of PLA probes) per cell were counted by semiautomated image analysis using ImageJ. n ≥ 34 cells scored in the experiment. Mean values and standard error of the mean (S.E.M) are represented (***p = 0.0001 for sT (upper panel), ***p = 0.0004 for cyclin E (lower panel), ****p < 0.0001). c Protein expression levels of PLA samples. Protein expression was evaluated by immunoblot analysis to validate successful transfection. Quantitative Infrared fluorescence immunoblotting was performed for T antigens, HA-FBW7 (WT and mut), c-Myc, Cyclin E, GFP and alpha-tubulin, respectively.