Some anaerobic microorganisms convert inorganic mercury (Hg) into the neurotoxin methylmercury, which can bioaccumulate and biomagnify in the food web. While the genetic basis of microbial mercury methylation is known, factors that control net methylmercury production in the environment are still poorly understood. Here, it is shown that mercury methylation can be substantially enhanced by one form of an exogenous copper-binding compound (methanobactin) produced by some methanotrophs, but not by another. This novel finding illustrates that complex interactions exist between microbes and that these interactions can potentially affect the net production of methylmercury in situ.
KEYWORDS: anaerobic bacteria, mercury methylation, methanotrophs, methylmercury
ABSTRACT
Microbial production of the neurotoxin methylmercury (MeHg) is a significant health and environmental concern, as it can bioaccumulate and biomagnify in the food web. A chalkophore or a copper-binding compound, termed methanobactin (MB), has been shown to form strong complexes with mercury [as Hg(II)] and also enables some methanotrophs to degrade MeHg. It is unknown, however, if Hg(II) binding with MB can also impede Hg(II) methylation by other microbes. Contrary to expectations, MB produced by the methanotroph Methylosinus trichosporium OB3b (OB3b-MB) enhanced the rate and efficiency of Hg(II) methylation more than that observed with thiol compounds (such as cysteine) by the mercury-methylating bacteria Desulfovibrio desulfuricans ND132 and Geobacter sulfurreducens PCA. Compared to no-MB controls, OB3b-MB decreased the rates of Hg(II) sorption and internalization, but increased methylation by 5- to 7-fold, suggesting that Hg(II) complexation with OB3b-MB facilitated exchange and internal transfer of Hg(II) to the HgcAB proteins required for methylation. Conversely, addition of excess amounts of OB3b-MB or a different form of MB from Methylocystis strain SB2 (SB2-MB) inhibited Hg(II) methylation, likely due to greater binding of Hg(II). Collectively, our results underscore the complex roles of microbial exogenous metal-scavenging compounds in controlling net production and bioaccumulation of MeHg in the environment.
IMPORTANCE Some anaerobic microorganisms convert inorganic mercury (Hg) into the neurotoxin methylmercury, which can bioaccumulate and biomagnify in the food web. While the genetic basis of microbial mercury methylation is known, factors that control net methylmercury production in the environment are still poorly understood. Here, it is shown that mercury methylation can be substantially enhanced by one form of an exogenous copper-binding compound (methanobactin) produced by some methanotrophs, but not by another. This novel finding illustrates that complex interactions exist between microbes and that these interactions can potentially affect the net production of methylmercury in situ.
INTRODUCTION
Microbial production of mono-methylmercury (MeHg or CH3Hg+), a potent neurotoxin, is a significant environmental and human health concern, as MeHg readily bioaccumulates and biomagnifies in the food web, particularly in fish and rice (1–3). A small group of anaerobic microorganisms possessing the gene pair hgcAB converts inorganic mercury [Hg(II)] to MeHg in aquatic and terrestrial environments (4–6). However, many environmental factors, such as the presence or absence of various organic/inorganic ligands, sulfides, and minerals, have been shown to significantly influence the rate and extent of Hg(II) methylation by these microbes (7–12). In particular, aqueous Hg(II) complexation with low-molecular-weight (LMW) thiols and naturally dissolved organic matter (DOM) has been shown to either increase or decrease Hg(II) methylation, depending on the chemical structure and concentration of the ligand, as well as the bacterial strain itself (9–11, 13). Specifically, most LMW thiols (e.g., cysteine, glutathione, and penicillamine) and DOM have been found to increase Hg(II) methylation by sulfate-reducing bacteria, such as Desulfovibrio desulfuricans ND132. On the other hand, only certain thiols (e.g., cysteine) can increase Hg(II) methylation by iron-reducing bacteria, such as Geobacter sulfurreducens PCA, whereas others (e.g., glutathione and penicillamine) inhibit this activity (9–11). These differences are partially explained by the varying abilities of these microbes to competitively take up Hg(II) that is complexed with the ligands in solution (14, 15).
Net MeHg production in the environment, however, also depends on biological degradation of MeHg (16, 17). Recent studies have discovered that some methanotrophs, e.g., Methylosinus trichosporium OB3b, can take up and degrade MeHg, and that this activity requires the expression or production of a copper-binding compound or chalkophore, called methanobactin (MB) (17). Methanobactin binds copper strongly, with measured binding constants ranging from 1018 to 1058 M−1, and is believed to be an important mechanism for copper uptake by methanotrophs in the environment (18–21). Although MB concentrations have never been reported in situ, concentrations of MB ranging from 10 to 50 μM are consistently observed in laboratory studies, suggesting that environmental concentrations of MB may be significant (19, 22).
Two general forms of MB have been described to date: group I MBs, such as MB from M. trichosporium OB3b (OB3b-MB), have two oxazolone rings with an internal disulfide bridge, whereas group II MBs, such as that made by Methylocystis strain SB2 (SB2-MB), contain one oxazolone ring and an imidazolone or pyrazinedione ring with an associated sulfate group. Not surprisingly, because of the N2S2 moieties used for copper binding, MBs are also shown to bind a variety of metal ions, including Hg(II), although the Hg(II) binding constant with SB2-MB (∼1025 M−1) is orders of magnitude higher than that with OB3b-MB (1.0 × 107 M−1) (23–25).
The formation of strong Hg(II)-MB complexes has been shown to alleviate Hg[II] toxicity to methanotrophs in laboratory experiments (24). It has also been shown that MB can affect the activity of denitrifying bacteria via copper sequestration (26). Little is known, however, as to whether MB affects the activity of other microbes, such as mercury-methylating bacteria. Does Hg(II) binding by MB impede or enhance microbial MeHg production? Such a question has environmental relevance, as methanotrophs thrive in a wide range of environments, including freshwater and marine sediments and wetlands (19, 27, 28), particularly at the oxic/anoxic interface. Evidence of the presence and activity of methanotrophs has also been reported in micro-oxic/anoxic conditions (29–31). Since MeHg is also produced at the top few centimeters of the anoxic interface (32, 33), MB may thus be present in these locations either due to diffusion/vertical transport or contiguous production. Therefore, the present study was carried out to address the following specific objectives: (i) determine the effect of MB on Hg(II) methylation rates by two mercury-methylating bacteria, D. desulfuricans ND132 and G. sulfurreducens PCA, which respond differently to various Hg(II) complexes of organic ligands, such as LMW thiols and DOM (9, 11, 34); and (ii) examine the influence of MB on Hg(II) sorption, internalization, and species distributions in these microorganisms.
RESULTS AND DISCUSSION
MB influences methylmercury production.
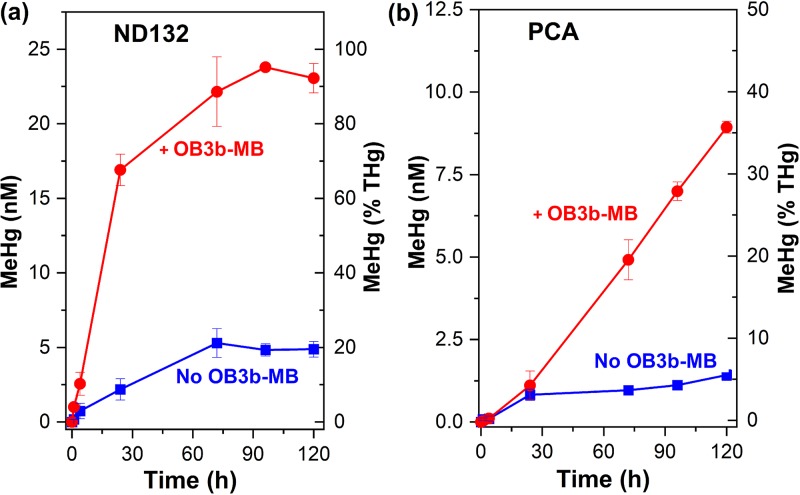
The addition of OB3b-MB increased MeHg production substantially by both D. desulfuricans ND132 and G. sulfurreducens PCA (Fig. 1). Without addition of OB3b-MB, D. desulfuricans ND132 and G. sulfurreducens PCA produced only 5 and 1.3 nM MeHg after 120 h [or ∼20% and 5% of Hg(II)], respectively, consistent with previous findings under similar experimental conditions (14, 35). When cells were incubated with preequilibrated Hg(II)-OB3b-MB complexes, however, MeHg production by D. desulfuricans ND132 increased sharply within 24 h and reached a plateau after ∼72 h (Fig. 1a). More than 90% of the added Hg(II) was methylated after 72 h (Fig. 1a), the highest methylation efficiency observed for this strain in phosphate-buffered saline (PBS) (13, 14, 35). Compared to assays in the absence of OB3b-MB, the methylation rate constant in the presence of 25 μM OB3b-MB increased more than an order of magnitude (from 3 × 10−3 to 3.9 × 10−2 h−1). Previous studies reported a methylation efficiency of 40 to 60% by D. desulfuricans ND132 in the presence of 50 to 100 μM thiols, such as cysteine and glutathione (13, 14, 36). Similarly, for G. sulfurreducens PCA, the presence of OB3b-MB increased the methylation rate nearly 6-fold, from 0.6 × 10−3 to 3.5 × 10−3 h−1, and resulted in >35% of Hg(II) methylated in 120 h (Fig. 1b). This enhanced methylation rate and extent was much higher than that observed in the presence of 50 μM cysteine, which increased Hg(II) methylation rates by 2- to 3-fold, compared to no-cysteine assays (10, 13, 37). Moreover, methylation in the presence of OB3b-MB continued to increase over 120 h (Fig. 1b), whereas previous studies have shown that Hg(II) methylation by this strain usually reaches a plateau after 24 to 48 h either in the presence or absence of cysteine (10, 13, 37).
FIG 1.
Effect of methanobactin (MB) from Methylosinus trichosporium OB3b (OB3b-MB) on methylmercury (MeHg) production by washed cells of D. desulfuricans ND132 (a) and G. sulfurreducens PCA (b) in phosphate-buffered saline (PBS). OB3b-MB (25 μM) was preequilibrated with 25 nM Hg(II) for 30 min prior to the addition of cells. Error bars represent one standard deviation (n = 4 to 6 replicate samples) from 3 independent batch experiments.
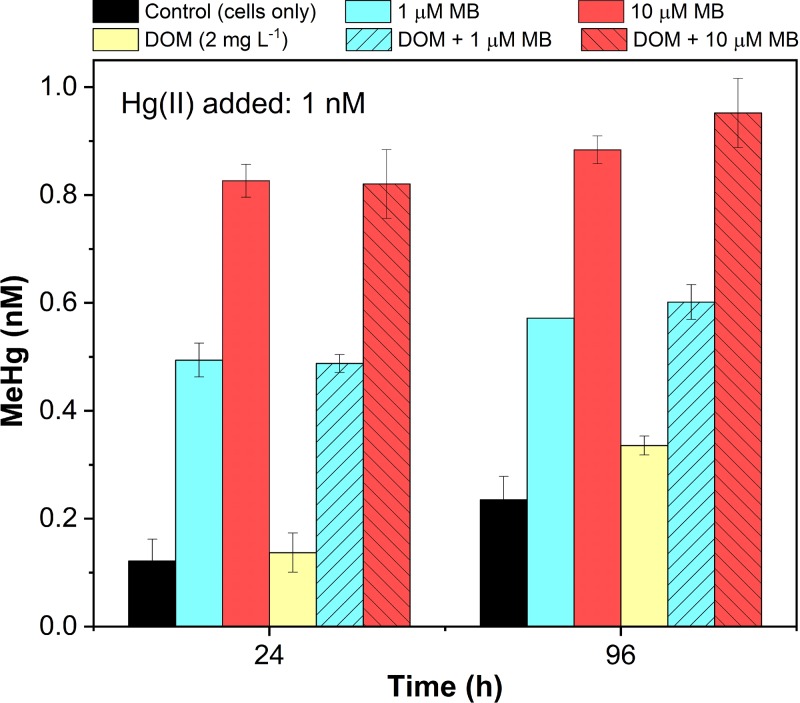
More significantly, the enhancement effect by OB3b-MB was also observed at low concentrations of Hg(II) (1 nM) and MB (1 and 10 μM) either in the presence or absence of a typical concentration of natural DOM (2 mg liter−1) (Fig. 2), which is known to bind Hg(II) and affect its uptake and methylation (11, 38). Compared to the absence of OB3b-MB, addition of only 1 μM OB3b-MB increased MeHg production by 4- to 5-fold in 24 h at the initial Hg(II) concentration of 1 nM by D. desulfuricans ND132, regardless of the presence or absence of DOM (Fig. 2). In the presence of DOM alone, MeHg production increased about 50%, as previously observed (11). However, addition of 10 μM OB3b-MB increased MeHg production 7- to 8-fold at 24 h, and nearly 90% of the added Hg(II) was methylated at 96 h. These results are comparable to those observed at relatively high concentrations of Hg(II) (25 nM) and OB3b-MB (25 μM) (Fig. 1a), although smaller amounts of OB3b-MB (1 to 10 μM) were required to cause the same fraction of Hg(II) (60 to 90%) to be methylated at a lower Hg(II) concentration (1 nM) (Fig. 2). These results demonstrate the potential role of the chalkophore methanobactin in enhancing MeHg production under environmentally relevant conditions, since DOM (containing various small thiols) showed little influence or only slightly increased Hg(II) methylation.
FIG 2.
Evaluation of DOM (2 mg liter−1) and low concentrations of Hg(II) (1 nM) and OB3b-methanobactin (MB, 1 or 10 μM) on methylmercury (MeHg) production by washed cells of D. desulfuricans ND132 (108 cells ml−1) in PBS. Hg(II) was preequilibrated with MB for 30 min prior to the addition of ND132 cells. Error bars represent the deviation between the two replicate samples (X1 and X2), defined as the absolute value of (X1 − X2)/2.
MB influences Hg(II) sorption, species distribution, and methylation.
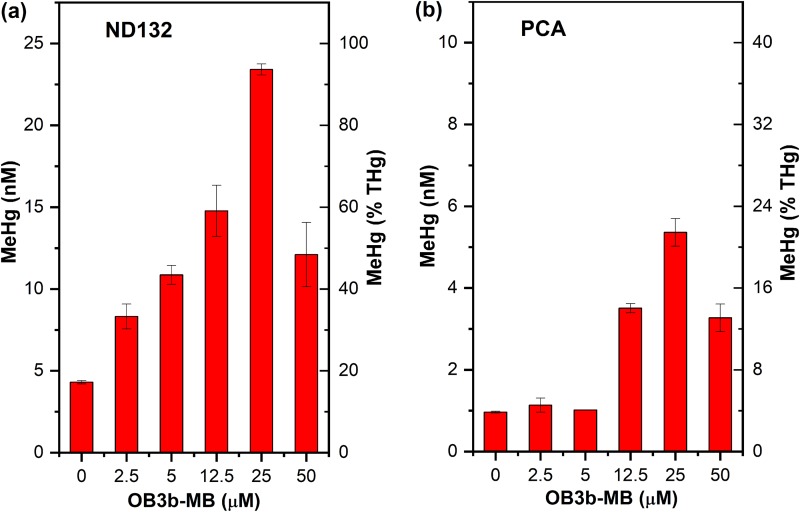
The magnitude of enhanced Hg(II) methylation by OB3b-MB is remarkable and unexpected, as MBs are known to form strong complexes with Hg(II) (23, 24). It was initially presumed that the presence of MB would inhibit Hg(II) methylation, particularly by G. sulfurreducens PCA, but the inverse was observed. To elucidate how OB3b-MB enhanced Hg(II) methylation by D. desulfuricans ND132 and G. sulfurreducens PCA, separate experiments were performed to examine the concentration effects of OB3b-MB on MeHg production and Hg(II) species distribution in these microbes. We found that MeHg production increased with increasing OB3b-MB concentrations up to 25 μM (Fig. 3). At this MB concentration, ∼90% and 20% of the added Hg(II) were converted to MeHg in 72 h by D. desulfuricans ND132 and G. sulfurreducens PCA, respectively. However, increasing OB3b-MB to 50 μM decreased Hg(II) methylation by both microbes, indicating that the presence of low to moderate concentrations of OB3b-MB favored the production of MeHg by both bacterial strains. This enhancement effect appears not to be due to altered microbial activity, as we found no significant changes in cell metabolic activity as assessed by measuring consumption rates of an added electron donor (pyruvate) and acceptor (fumarate), or the production rates of acetate and succinate either with or without addition of OB3b-MB by D. desulfuricans ND132 (Fig. S1 in the supplemental material). We also found no significant degradation of OB3b-MB over time, as measured by UV-Vis and liquid 1H-NMR spectroscopy (Fig. S2). While we cannot unequivocally exclude the possibility of altered metabolic activity, we hypothesized that low to moderate concentrations of OB3b-MB likely increased the amount of available Hg(II) by forming Hg(II)-OB3b-MB complexes or by inhibiting Hg(II) from sorbing onto some unknown binding sites that caused Hg(II) immobilization or prevented it from being methylated, as previously observed (13, 14, 39). However, a high MB concentration may increase competition between OB3b-MB and cells for Hg(II) binding, resulting in decreasing Hg(II) uptake and methylation. In the case of G. sulfurreducens PCA, the presence of OB3b-MB could also inhibit Hg(II) reduction (described below) (10, 40, 41), thereby increasing available Hg(II) for methylation.
FIG 3.
Effect of various OB3b-MB concentrations on methylmercury (MeHg) production by washed cells of D. desulfuricans ND132 (a) and G. sulfurreducens PCA (b) at 72 h in phosphate-buffered saline (PBS). Hg(II) (25 nM) was preequilibrated with the indicated amounts of OB3b-MB for 30 min prior to addition of cells. Error bars represent the deviation between the two replicate samples (X1 and X2), defined as the absolute value of (X1 − X2)/2.
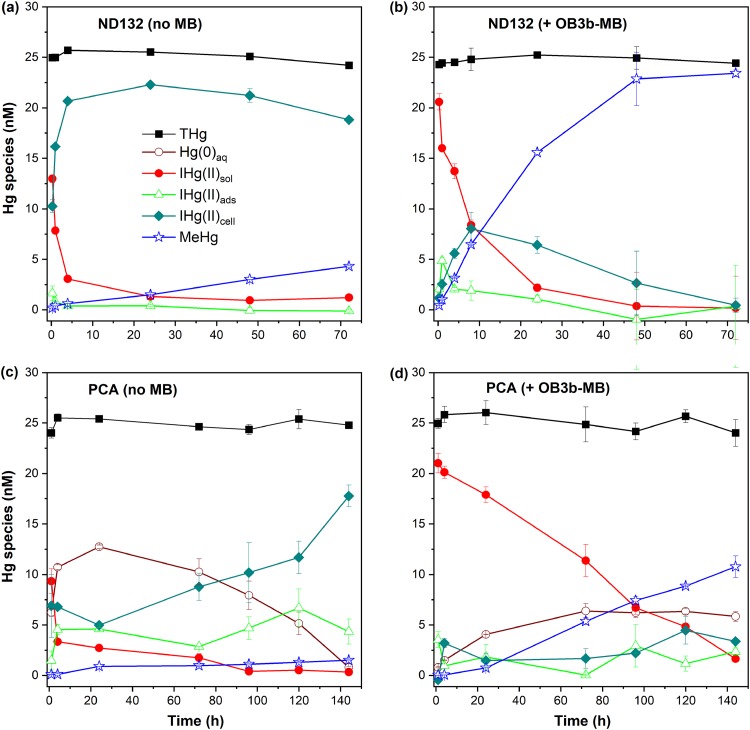
These hypotheses are supported by analyses of Hg(II) species distributions in the absence or presence of OB3b-MB during methylation assays (Fig. 4). Although MeHg distributions were also determined (Fig. S3), here we focus only on the adsorption and uptake of the inorganic Hg(II) species, as most MeHg was rapidly excreted in solution, as reported elsewhere (34, 39). Without OB3b-MB, most of the added Hg(II) (21 nM or ∼84%) was rapidly taken up by D. desulfuricans ND132 (Fig. 4a). A small amount of Hg(II) was converted to MeHg (∼0.4 nM at 4 h and 4.3 nM at 72 h). In the presence of 25 μM OB3b-MB, a substantially larger amount (∼14 nM) of Hg(II) remained in solution at 4 h (Fig. 4b) compared to that in the absence of OB3b-MB (∼3 nM; Fig. 4a). Additionally, the solution-phase Hg(II) concentration decreased gradually over time and coincided with increasing MeHg production in the presence of OB3b-MB (Fig. 4b). These results suggest that OB3b-MB formed soluble complexes with Hg(II) and either prevented it from rapidly sorbing to the cell surface and/or becoming immobilized by cellular material.
FIG 4.
Mercury (Hg) species distributions during Hg(II) methylation assays with washed cells of D. desulfuricans ND132 (a, b) and G. sulfurreducens PCA (c, d) in phosphate-buffered saline (PBS). Experiments in panels b and d were performed in the presence of 25 μM OB3b-MB, which was preequilibrated with 25 nM Hg(II) in PBS prior to the addition of cells (108 cells ml−1). THg, Hg(0)aq, IHg(II)sol, IHg(II)ads, IHg(II)cell, and MeHg denote concentrations of total Hg, dissolved elemental Hg(0), soluble inorganic Hg(II), cell-surface adsorbed Hg(II), intracellular uptake of Hg(II), and total methylmercury (MeHg), respectively. Error bars represent the deviation between the two replicate samples (X1 and X2), defined as the absolute value of (X1 − X2)/2.
Similar to that observed with D. desulfuricans ND132, Hg(II) was mostly taken up or internalized within a few hours of incubation with G. sulfurreducens PCA in the absence of OB3b-MB, with negligible amounts of Hg(II) left in solution after 96 h (Fig. 4c). Only a small amount of Hg(II) was methylated (∼1.3 nM). In the presence of OB3b-MB, however, MeHg production increased substantially to ∼7 nM at 96 h (Fig. 4d), as noted above. Meanwhile, a much larger amount (∼71%) of Hg(II) remained in solution at 24 h (Fig. 4d), compared to ∼10% of Hg(II) in the absence of OB3b-MB. About 7 nM (or 28%) of the added Hg(II) remained in solution after 96 h in the presence of OB3b-MB (Fig. 4d), compared to <2% in the absence of OB3b-MB (Fig. 4c). We also observed a smaller amount of Hg(II) sorption on G. sulfurreducens PCA but a much larger amount of Hg(II) methylation (∼10 nM) in the presence than absence of OB3b-MB. Additionally, the presence of OB3b-MB substantially decreased Hg(II) reduction within the first 24 to 48 h (Fig. 4d). A much smaller amount (∼4 nM at 24 h) of Hg(II) was reduced due to complexation between Hg(II) and OB3b-MB. Without OB3b-MB, approximately half of the added Hg(II) was reduced in 24 h, although it was reoxidized over time (Fig. 4c), as G. sulfurreducens PCA is known to reduce or oxidize Hg over time (10, 40).
Analyses of Hg(II) species distributions for both D. desulfuricans ND132 and G. sulfurreducens PCA indicate that, in the absence of OB3b-MB, Hg(II) internalization was rapid, yet Hg methylation rates were much lower than when OB3b-MB was present. Inversely, in the presence of OB3b-MB, Hg(II) sorption and internalization were lower and more Hg(II) remained in solution, but much larger amounts of Hg(II) were methylated. These observations suggest that Hg(II) internalization does not by itself lead to Hg(II) methylation, as observed in previous studies (13, 14, 39). The data indicate that the mechanism of Hg(II) uptake may control internal transfer of Hg(II) to the HgcAB proteins responsible for its methylation. In the presence of OB3b-MB, Hg(II) sorption and uptake via more predominant, lower-affinity binding sites not involved in transfer of Hg(II) to HgcAB were likely prevented, thus facilitating more direct transfer of Hg(II) to HgcAB. This phenomenon could explain the observed lower rates of Hg(II) sorption and uptake in the presence of OB3b-MB. This hypothesis is supported by the finding that, at best, only slightly enhanced Hg(II) methylation (<15%) was observed when D. desulfuricans ND132 was exposed to Hg(II) for 1 h prior to the addition of OB3b-MB (Fig. S4). In this case, most Hg(II) would be rapidly sorbed or immobilized, as observed in the absence of OB3b-MB (Fig. 4a), and thus become unavailable for methylation.
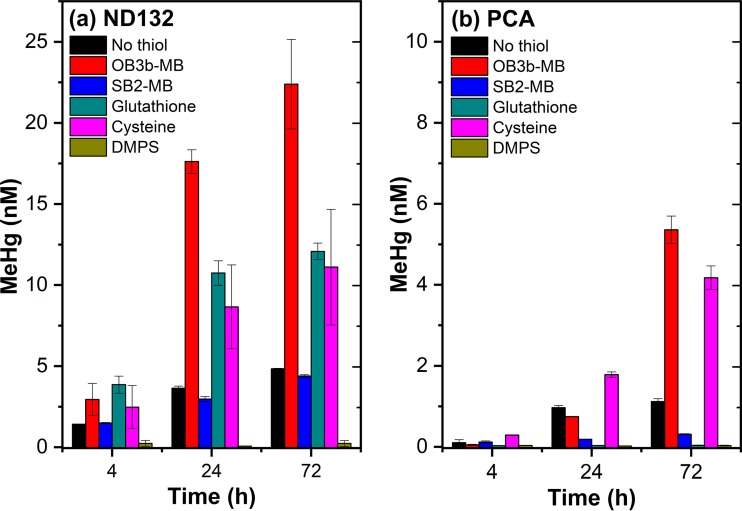
Comparisons between MBs and thiols on methylmercury production.
To further explore mechanisms of MB-enhanced Hg(II) methylation, we compared the effect of OB3b-MB with a structurally different methanobactin, SB2-MB (Table S1), and several LMW thiols (cysteine, glutathione [GSH], and 2,3-dimercapto-1-propanesulfonic acid [DMPS]). These thiols were chosen because of known impacts on Hg(II) methylation by D. desulfuricans ND132 and G. sulfurreducens PCA (9, 10, 13). Interestingly, despite its similarities with OB3b-MB, SB2-MB did not stimulate Hg(II) methylation by D. desulfuricans ND132 (Fig. 5a) and inhibited methylation by G. sulfurreducens PCA (Fig. 5b). More specifically, D. desulfuricans ND132 produced only about 3.0 and 4.4 nM MeHg at 24 and 72 h, respectively, in the presence of SB2-MB, which are levels similar to those produced in the absence of either OB3b-MB, SB2-MB, or thiols. For G. sulfurreducens PCA, the presence of SB2-MB nearly abolished MeHg production, with only 0.3 nM MeHg produced in 72 h (Fig. 5b).
FIG 5.
Comparisons between LMW thiols and OB3b-MB and SB2-MB on methylmercury (MeHg) production by washed cells (108 ml−1) of D. desulfuricans ND132 (a) and G. sulfurreducens PCA (b) in phosphate-buffered saline (PBS). The added Hg(II) concentration was 25 nM and MB concentration was 25 μM. The added thiol concentration was 50 μM for glutathione and cysteine and 100 μM for 2,3-dimercapto-1-propanesulfonic acid (DMPS), as previously described (13, 14). Error bars represent one standard deviation (n = 4 replicate samples) from 2 independent batch experiments.
Consistent with previous observations (10, 11, 13), addition of cysteine and glutathione (50 μM each) increased Hg(II) methylation by D. desulfuricans ND132, albeit to lower levels than those observed in the presence of OB3b-MB (Fig. 5a). After 72 h, about 11 to 12 nM MeHg was produced by D. desulfuricans ND132 in the presence of cysteine or glutathione compared to 22.4 nM MeHg in the presence of OB3b-MB. Addition of cysteine also enhanced MeHg production by G. sulfurreducens PCA [4.1 nM or 16% of Hg(II) after 72 h], but glutathione halted Hg(II) methylation (Fig. 5b) (9, 10). Similarly, the di-thiolate chelator DMPS (100 μM) completely inhibited Hg(II) methylation by both D. desulfuricans ND132 and G. sulfurreducens PCA, as reported previously (13, 14). Collectively, these results indicate that, similar to the effects of many thiol compounds, methylating bacteria respond differently to different methanobactins. Such a differential response may be due to physiological differences between sulfate and iron reducers, e.g., different surface-binding moieties (34, 42) and/or uptake mechanisms (9, 11, 14). SB2-MB behaved somewhat similarly to glutathione or DMPS in affecting Hg(II) methylation, whereas OB3b-MB behaved like cysteine in enhancing Hg(II) methylation.
These observations imply that the thermodynamic stability of Hg(II)-MB complexes and their molecular structures may be important in controlling Hg(II) interactions and uptake by D. desulfuricans ND132 and G. sulfurreducens PCA. Previous studies have shown that SB2-MB, which contains one oxazolone and one imidazolone ring (Table S1), preferentially binds Hg(II) over Cu(II), whereas OB3b-MB has two oxazolone rings and preferentially binds Cu(II) over Hg(II) (23, 25). Both forms of MB use N2S2 moieties on these heterocyclic rings to bind Hg(II), but the binding constant of SB2-MB for Hg(II) (∼1025 mol−1) is about 18 orders of magnitude higher than that of OB3b-MB (1.0 × 107 mol−1) (23–25), which could thus slow down or prevent Hg(II) from sorption or uptake, as observed with OB3b-MB (Fig. 4). The difference in Hg(II) binding affinity could also explain why increasing OB3b-MB concentrations (i.e., 50 μM) decreased MeHg production (Fig. 3), as there was increased competition between OB3b-MB and cells for Hg(II) binding.
Cell surface structures and binding sites could also be crucial in affecting Hg(II) sorption and ligand-exchange reactions. Only certain binding sites may be responsible for exchanging Hg(II) and directing it to the HgcAB proteins for methylation, whereas others sorb Hg(II) and immobilize it, making it unavailable for methylation. The complexation of Hg(II) by OB3b-MB likely facilitated Hg(II) to those binding sites where exchange and transfer reactions took place. The observation that OB3b-MB enhanced Hg(II) methylation more than cysteine did support the notion that Hg(II) complexed by OB3b-MB may be more readily available and transferred to HgcAB than that complexed by cysteine. Additionally, relative abundances and steric arrangements of cell binding sites may partially explain differences observed in Hg(II) methylation between the two bacterial strains, as D. desulfuricans ND132 cells are known to contain more sulfhydryl functional groups than G. sulfurreducens PCA cells (11, 42).
Hg(II) is unlikely to be taken up as an intact Hg(II)-OB3B-MB complex based on the theory of bio-uptake of metal ions (43, 44). That is, OB3b-MB is a ribosomally synthesized posttranslationally modified peptide with molecular weight of 1,154.3 Da, and its uptake requires a specific TonB-dependent transporter, MbnT (45). A simple blastP analysis indicates that there is no gene in the genome of D. desulfuricans ND132 encoding a product similar to MbnT, and possibly two genes in the genome of G. sulfurreducens PCA. However, the E values for these two genes are quite high (>10−4), indicating that these TonB-dependent transporters are dissimilar to MbnT and thus unlikely to take up metal-MB complexes. Additional studies are thus warranted to verify whether MB can be taken up by these microbes. The postulation that MB is not taken up by D. desulfuricans ND132 and G. sulfurreducens PCA, however, is indirectly supported by the observation that little or no enhanced Hg(II) methylation was observed when the order-of-addition sequence for OB3b-MB and ND132 was reversed (Fig. S3), i.e., cells were incubated with Hg(II) for 1 h before addition of OB3b-MB. Only when Hg(II) and OB3b-MB were preequilibrated was significant enhancement of Hg(II) methylation observed. On the contrary, a previous study has shown that cysteine could enhance Hg(II) methylation, regardless of whether cysteine is added before or after D. desulfuricans ND132 has reacted with Hg(II) (13). This suggests that cysteine, but not OB3b-MB, may be taken up by D. desulfuricans ND132 and thus make some immobilized Hg(II) available for methylation. Other studies have also shown that cysteine could be degraded to form sulfide by both D. desulfuricans ND132 and G. sulfurreducens PCA and could thus decrease Hg(II) availability by forming HgS (11, 46).
In summary, we report that OB3b-MB increases the rate and extent of Hg(II) methylation more than that observed in the presence of LMW thiols and DOM (10, 11, 13, 39). This enhanced Hg(II) methylation is hypothesized to result from Hg(II) complexation with OB3b-MB (23–25), which facilitates Hg(II) exchange with the cell and then transfer to HgcAB for methylation. This hypothesis is supported by Hg(II) speciation analyses (Fig. 3), in which the rates of Hg(II) sorption and internalization were inhibited but methylation increased in the presence of OB3b-MB. Without OB3b-MB, the sorption and internalization of Hg(II) were rapid, but the rate and extent of Hg(II) methylation were much lower than those levels observed in the presence of OB3b-MB.
Methanobactins, however, vary in forming complexes with Hg(II) and affecting Hg(II) uptake and methylation, as observed with various thiol compounds (9–11, 13). SB2-MB showed little influence on Hg(II) methylation by D. desulfuricans ND132 and inhibited Hg(II) methylation by D. desulfuricans PCA, suggesting that structural differences between SB2-MB and OB3b-MB played an important role in cell exchange and uptake of Hg(II). The Hg(II)-SB2-MB complex may be too strong or sterically hindered for cells to compete with the ligand for Hg(II) binding or uptake (Fig. 4), whereas Hg(II)-OB3b-MB and Hg(II)-cysteine complexes facilitate Hg(II) exchange and its transfer to HgcAB. A high ligand concentration (e.g., OB3b-MB, cysteine, etc.) can increase competition for Hg(II) binding, leading to decreased methylation. Therefore, structural differences and thermodynamic stability of Hg(II)-ligand complexes appear to control the ability of cells to competitively exchange Hg(II) and then direct it to the HgcAB for methylation.
The present study suggests the potential role of MBs in affecting microbial methylation in the environment. As stated above, not only do methanotrophs predominate at the oxic/anoxic interface where CH4 and MeHg may be observed, their presence/activity is measurable under micro-oxic/anoxic conditions (29–31). Further, methanotrophs have been observed to secrete substantial (∼50 μM) amounts of MB in laboratory systems (19, 27, 28), and in vitro studies show that methanotrophs, through production of MB, can affect the activity of denitrifying bacteria (26). Although there are no published studies that have directly examined either the amount or location of MB produced in situ, such production likely exists and may thus influence Hg(II) methylation by organisms such as D. desulfuricans ND132 and G. sulfurreducens PCA near the anoxic interface. Interestingly, however, methanotrophs can also degrade MeHg, and MB is necessary for demethylation to occur (17). Therefore, the production, release, and transport of exogenous MBs could significantly impact both the production and degradation of MeHg. Future studies are warranted to further explore potential interactions among various microbial communities and complex interplay among various MBs and LMW thiols in controlling Hg(II) chemical speciation, microbial uptake, and ultimately net MeHg production in natural environments.
MATERIALS AND METHODS
Reagents and chemical preparation.
Reagent-grade organic ligands, including l-cysteine (CYS), glutathione (GSH), and 2,3-dimercapto-1-propanesulfonic acid (DMPS), were purchased from Sigma-Aldrich. OB3b-MB and SB2-MB were isolated and characterized, as described in detail elsewhere (22–24), and their chemical structures are shown in Table S1 of the supplemental material. Phosphate-buffered saline (PBS), consisting of 0.14 M NaCl, 3 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4 at pH 7.4, was sterilized via autoclaving and then deoxygenated by boiling and purging with ultrahigh purity N2 gas for at least 2 h (10, 11, 47). Stock solutions of PBS were subsequently kept in an anoxic glove chamber (with 98% N2 and 2% H2) for at least 24 h before use. The MB solution was prepared freshly by dissolving a preweighed amount of MB in PBS in a glass vial and then diluted and used for Hg(II) methylation assays.
Microbial Hg(II) methylation and uptake assays.
The sulfate-reducing bacterium D. desulfuricans ND132 and the iron-reducing bacterium G. sulfurreducens PCA were used as two model Hg(II) methylators to assess the impact of MB on microbial Hg(II) methylation (9, 36, 37). D. desulfuricans ND132 was cultured in a modified minimal organic yeast (MOY) medium containing 40 mM fumarate and 40 mM pyruvate, whereas G. sulfurreducens PCA was cultured in the nutrient broth Basal salts containing 40 mM fumarate and 20 mM acetate at 30°C, as previously described (10, 11, 47). Cells were harvested during mid-exponential phase at an optical density at 600 nm (OD600) of 0.3 and washed three times by repeated centrifugation (at 1,200 × g, 10 min, 25°C) and resuspension in PBS. All washing steps and subsequent Hg(II) methylation assays were conducted in the anoxic chamber.
Hg(II) methylation assays were performed by preequilibrating a solution with fixed Hg(II) (25 nM as HgCl2) and MB (25 μM) concentrations for 30 min in PBS in an anoxic chamber. An aliquot of the washed cell suspension (0.5 ml) was then added to give a final concentration of 1 × 108 cells ml−1 in 4-ml amber glass vials. Samples were supplemented once (at t = 0 h) with 1 mM (each) pyruvate and fumarate for D. desulfuricans ND132 or 1 mM (each) acetate and fumarate for G. sulfurreducens PCA as the respective electron donor and acceptor. Final solution volume was 1 ml. All vials were immediately sealed with polytetrafluoroethylene (PTFE)-lined silicone screw caps and placed on a rotary shaker at 90 rpm in the anoxic chamber in the dark at 23°C. At select time points, replicate samples (2–4) were taken out of the anoxic chamber and an aliquot (0.1 ml) was immediately extracted and analyzed for MeHg (described below). The remaining sample was oxidized in BrCl (5% vol/vol)-HCl (0.2 M) overnight at 4°C for total Hg analysis. Similar experiments were performed in the presence of various concentrations of MB (2.5 to 50 μM), cysteine, glutathione, or DMPS, representing important thiol ligands commonly observed and/or used in biogeochemical investigations of Hg(II) transformations (9, 13, 14). Additional methylation assays were performed at low Hg(II) (1 nM) and MB (1 and 10 μM) concentrations either in the absence or presence of DOM (2 mg liter−1), as previously described (11, 48, 49). Parallel experiments were also performed without cells as negative controls.
To determine Hg(II) uptake and species distribution during Hg(II) methylation, a strong Hg(II)-chelator, DMPS, was added to a subset of sample vials to wash off cell-surface-adsorbed Hg(II) and MeHg, as previously described (13, 14, 47). Briefly, a set of six sample vials was taken out of the anoxic chamber and analyzed for both inorganic Hg(II) and MeHg species as follows. Two of the samples were used for MeHg and total Hg(II) (THg) analyses, and another two samples were filtered through 0.2-μm Acrodisc syringe filters (13 mm, Pall) to remove cells and analyzed for total soluble Hg(II) (Hgsol) and MeHgsol in the same manner. Soluble inorganic Hg(II) [IHg(II)sol] was calculated by subtracting MeHgsol from Hgsol. A small aliquot (10 μl) of DMPS was added to the two remaining samples to obtain a final concentration of 0.1 mM. These samples were equilibrated for an additional 15 min to wash off cell-surface-adsorbed Hg(II) (Hgads) (including the adsorbed MeHg, or MeHgads), as DMPS forms strong complexes with Hg(II) (13, 14, 47). Samples were again filtered and analyzed in the same manner. Hgads was determined by the difference between the amounts of Hg(II) in filtrate solutions with and without the DMPS wash. Similarly, surface-adsorbed inorganic Hg(II) [IHg(II)ads] was calculated by subtracting MeHgads from Hgads, and intracellular Hg(II) [IHg(II)cell] was calculated by subtracting Hgsol and Hgads from THg [i.e., IHg(II)cell = THg − Hgsol − Hgads] (13, 14). MeHg distributions were also calculated in the same manner.
Hg(II) and MeHg analyses.
Total Hg(II) was determined by reduction with SnCl2 and detection of purgeable gaseous Hg(0) using a cold vapor atomic absorption spectrometer (CVAAS) (RA-915+, Ohio Lumex Co., Cleveland, OH) (10, 14, 38). The detection limit was 2.5 × 10−4 nM. MeHg was analyzed using a modified version of EPA Method 1630, where MeHg was extracted from samples via distillation and ethylation. Corrections for potential matrix interferences were made by adding isotopically labeled Me200Hg to each sample as an internal standard (13, 14, 35). Extracted MeHg was analyzed using an automated MERX purge and trap system (Brooks Rand, Seattle, WA), followed by detection on an inductively coupled plasma mass spectrometer (ICP-MS) (Elan DRC-e; PerkinElmer, Shelton, CT). One blank (Milli-Q water) and one randomly selected sample spiked with a known amount of MeHg were run in parallel every 30 samples. Additionally, MeHg reference standards (Brooks Rand) were run every 10 to 15 samples for quality assurance and quality control. The recovery of spiked MeHg standards was within 100% ± 10%, and the detection limit was about 3 × 10−5 nM MeHg. Data points in all figures represent an average of 2 to 6 replicate samples from 1 to 3 batch experiments, and error bars represent one standard deviation (n > 2) or the deviation between the two replicate samples (X1 and X2), defined as the absolute value of (X1 − X2)/2. Where error bars are not visible, the symbol size is greater than the magnitude of the deviation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiangping Yin for assistance in mercury and methylmercury analysis.
This research was supported in part by the Office of Biological and Environmental Research within the Office of Science of the U.S. Department of Energy (DOE) (grant number DE-SC0018059) and the U.S. National Science Foundation (grant numbers 1724430 and 1724744). X.Y. and L.W. were supported by the National Natural Science Foundation of China (NSFC) (grant number 41671485), the Natural Science Foundation of Shandong Province, China (grant number ZR2017MD008), and the Chinese Scholarship Council (CSC). Oak Ridge National Laboratory is managed by UT-Battelle, LLC, under contract number DE-AC05-00OR22725 with the DOE.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mason RP, Reinfelder JR, Morel FMM. 1995. Bioaccumulation of mercury and methylmercury. Water Air Soil Pollut 80:915–921. doi: 10.1007/BF01189744. [DOI] [Google Scholar]
- 2.Baumann Z, Mason RP, Conover DO, Balcom P, Chen CY, Buckman KL, Fisher NS, Baumann H. 2017. Mercury bioaccumulation increases with latitude in a coastal marine fish (Atlantic silverside, Menidia menidia). Can J Fish Aquat Sci 74:1009–1015. doi: 10.1139/cjfas-2016-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng XB, Li P, Qiu GL, Wang S, Li GH, Shang LH, Meng B, Jiang HM, Bai WY, Li ZG, Fu XW. 2008. Human exposure to methylmercury through rice intake in mercury mining areas, Guizhou Province, China. Environ Sci Technol 42:326–332. doi: 10.1021/es071948x. [DOI] [PubMed] [Google Scholar]
- 4.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. 2013. The genetic basis for bacterial mercury methylation. Science 339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 5.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, Johs A, Hurt RA, Bailey KL, Elias DA. 2013. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol 47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 6.Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, Palumbo AV, Somenahally AC, Elias DA. 2015. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 1:e1500675. doi: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benoit JM, Gilmour CC, Heyes A Mason RP, Miller CL. 2003. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems ACS Symp Ser 835:262–297. doi: 10.1021/bk-2003-0835.ch019. [DOI] [Google Scholar]
- 8.Jonsson S, Skyllberg U, Nilsson MB, Lundberg E, Andersson A, Bjorn E. 2014. Differentiated availability of geochemical mercury pools controls methylmercury levels in estuarine sediment and biota. Nat Commun 5:4624. doi: 10.1038/ncomms5624. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer JK, Rocks SS, Zheng W, Liang L, Gu B, Morel FMM. 2011. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proc Natl Acad Sci U S A 108:8714–8719. doi: 10.1073/pnas.1105781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, Morrell-Falvey JL, Rao B, Liang L, Gu B. 2014. Coupled mercury-cell sorption, reduction, and oxidation affecting methylmercury production by Geobacter sulfurreducens PCA. Environ Sci Technol 48:11969–11976. doi: 10.1021/es502537a. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Chen H, Lu X, Lin H, Christensen GA, Pierce EM, Gu B. 2017. Contrasting effects of dissolved organic matter on mercury methylation by Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132. Environ Sci Technol 51:10468–10475. doi: 10.1021/acs.est.7b02518. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Wu S, Zhao L, Lu X, Pierce EM, Gu B. 2019. Mercury sorption and desorption on organo-mineral particulates as a source for microbial methylation. Environ Sci Technol 53:2426–2433. doi: 10.1021/acs.est.8b06020. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y-R, Lu X, Zhao L, An J, He J-Z, Pierce EM, Johs A, Gu B. 2016. Effects of cellular sorption on mercury bioavailability and methylmercury production by Desulfovibrio desulfuricans ND132. Environ Sci Technol 50:13335–13341. doi: 10.1021/acs.est.6b04041. [DOI] [PubMed] [Google Scholar]
- 14.An J, Zhang L, Lu X, Pelletier DA, Pierce EM, Johs A, Parks JM, Gu B. 2019. Mercury uptake by Desulfovibrio desulfuricans ND132: passive or active? Environ Sci Technol 53:6264−6272. doi: 10.1021/acs.est.9b00047. [DOI] [PubMed] [Google Scholar]
- 15.Adediran G, Liem-Nguyen V, Song Y, Schaefer JK, Skyllberg U, Björn E. 2019. Microbial biosynthesis of thiol compounds: implications for speciation, cellular uptake and methylation of Hg(II). Environ Sci Technol 53:8187–8196. doi: 10.1021/acs.est.9b01502. [DOI] [PubMed] [Google Scholar]
- 16.Barkay T, Miller SM, Summers AO. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Gu WY, Zhao L, Ul Haque MF, DiSpirito AA, Semrau JD, Gu B. 2017. Methylmercury uptake and degradation by methanotrophs. Sci Adv 3:e1700041. doi: 10.1126/sciadv.1700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiSpirito AA, Semrau JD, Murrell JC, Gallagher WH, Dennison C, Vuilleumier S. 2016. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol Mol Biol Rev 80:387–409. doi: 10.1128/MMBR.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 20.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NL, Vorobev A, Haft DH, Vuilleumier S, Murrell JC. 2013. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ Microbiol 15:3077–3086. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 21.Semrau JD, DiSpirito AA, Gu WY, Yoon S. 2018. Metals and methanotrophy. Appl Environ Microbiol 84:e02289–17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, Gilles VS, DiSpirito AA. 2011. Isolation of methanobactin from the spent media of methane-oxidizing bacteria. Methods Enzymol 495:259–269. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 23.Baral BS, Bandow NL, Vorobev A, Freemeier BC, Bergman BH, Herdendorf TJ, Fuentes N, Ellias L, Turpin E, Semrau JD, DiSpirito AA. 2014. Mercury binding by methanobactin from Methylocystis strain SB2. J Inorg Biochem 141:161–169. doi: 10.1016/j.jinorgbio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Vorobev A, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, Bergman BH, Bandow NL, Semrau JD. 2013. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl Environ Microbiol 79:5918–5926. doi: 10.1128/AEM.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi DW, Do YS, Zea CJ, McEllistrem MT, Lee SW, Semrau JD, Pohl NL, Kisting CJ, Scardino LL, Hartsel SC, Boyd ES, Geesey GG, Riedel TP, Shafe PH, Kranski KA, Tritsch JR, Antholine WE, DiSpirito AA. 2006. Spectral and thermodynamic properties of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV), and Zn(II) binding by methanobactin from Methylosinus trichosporium OB3b. J Inorg Biochem 100:2150–2161. doi: 10.1016/j.jinorgbio.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 26.Chang J, Gu WY, Park D, Semrau JD, DiSpirito AA, Yoon S. 2018. Methanobactin from Methylosinus trichosporium OB3b inhibits N2O reduction in denitrifiers. ISME J 12:2086–2089. doi: 10.1038/s41396-017-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland NK, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 28.Knief C. 2015. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6:1346. doi: 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa RB, Okada DY, Delforno TP, Foresti E. 2019. Methane-oxidizing archaea, aerobic methanotrophs and nitrifiers coexist with methane as the sole carbon source. Int Biodeter Biodegr 138:57–62. doi: 10.1016/j.ibiod.2019.01.005. [DOI] [Google Scholar]
- 30.Kalyuzhnaya MG, Yang S Rozova ON, Smalley NE, Clubb J, Lamb A, Gowda GAN, Raftery D, Fu Y, Bringel F, Vuilleumier S, Beck DAC, Trotsenko YA, Khmelenina VN, Lidstrom ME. 2013. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 31.Lee HJ, Jeong SE, Kim PJ, Madsen EL, Jeon CO. 2015. High resolution depth distribution of Bacteria, Archaea, methanotrophs, and methanogens in the bulk and rhizosphere soils of a flooded rice paddy. Front Microbiol 6:639. doi: 10.3389/fmicb.2015.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris RC, Rudd JWM, Amyot M, Babiarz CL, Beaty KG, Blanchfield PJ, Bodaly RA, Branfireun BA, Gilmour CC, Graydon JA, Heyes A, Hintelmann H, Hurley JP, Kelly CA, Krabbenhoft DP, Lindberg SE, Mason RP, Paterson MJ, Podemski CL, Robinson A, Sandilands KA, Southworth GR, Louis VLS, Tate MT. 2007. Whole-ecosystem study shows rapid fish-mercury response to changes in mercury deposition. Proc Natl Acad Sci U S A 104:16586–16591. doi: 10.1073/pnas.0704186104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilmour CC, Henry EA, Mitchell R. 1992. Sulfate stimulation of mercury methylation in fresh-water sediments. Environ Sci Technol 26:2281–2287. doi: 10.1021/es00035a029. [DOI] [Google Scholar]
- 34.Lin H, Lu X, Liang L, Gu B. 2015. Thiol-facilitated cell export and desorption of methylmercury by anaerobic bacteria. Environ Sci Technol Lett 2:292–296. doi: 10.1021/acs.estlett.5b00209. [DOI] [Google Scholar]
- 35.Zhao L, Li Y, Zhang L, Zheng J, Pierce EM, Gu B. 2019. Mercury adsorption on minerals and its effect on microbial methylation. ACS Earth Space Chem 3:1338–1345. doi: 10.1021/acsearthspacechem.9b00039. [DOI] [Google Scholar]
- 36.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, Wall JD. 2011. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl Environ Microbiol 77:3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H, Lin H, Zheng W, Tomanicek SJ, Johs A, Feng X, Elias DA, Liang L, Gu B. 2013. Oxidation and methylation of dissolved elemental mercury by anaerobic bacteria. Nat Geosci 6:751–754. doi: 10.1038/ngeo1894. [DOI] [Google Scholar]
- 38.Gu B, Bian Y, Miller CL, Dong W, Jiang X, Liang L. 2011. Mercury reduction and complexation by natural organic matter in anoxic environments. Proc Natl Acad Sci U S A 108:1479–1483. doi: 10.1073/pnas.1008747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham AM, Bullock AL, Maizel AC, Elias DA, Gilmour CC. 2012. Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species. Appl Environ Microbiol 78:7337–7346. doi: 10.1128/AEM.01792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu H, Lin H, Zheng W, Rao B, Feng X, Liang L, Elias DA, Gu B. 2013. Mercury reduction and cell-surface adsorption by Geobacter sulfurreducens PCA. Environ Sci Technol 47:10922–10930. doi: 10.1021/es400527m. [DOI] [PubMed] [Google Scholar]
- 41.Lin H, Lu X, Liang L, Gu B. 2015. Cysteine inhibits mercury methylation by Geobacter sulfurreducens PCA mutant ΔomcBESTZ. Environ Sci Technol Lett 2:144–148. doi: 10.1021/acs.estlett.5b00068. [DOI] [Google Scholar]
- 42.Wang YW, Schaefer JK, Mishra B, Yee N. 2016. Intracellular Hg(0) oxidation in Desulfovibrio desulfuricans ND132. Environ Sci Technol 50:11049–11056. doi: 10.1021/acs.est.6b03299. [DOI] [PubMed] [Google Scholar]
- 43.Fein JB. 2017. Advanced biotic ligand models: using surface complexation modeling to quantify metal bioavailability to bacteria in geologic systems. Chem Geol 464:127–136. doi: 10.1016/j.chemgeo.2016.10.001. [DOI] [Google Scholar]
- 44.Slaveykova VI, Wilkinson KJ. 2005. Predicting the bioavailability of metals and metal complexes: critical review of the biotic ligand model. Environ Chem 2:9–24. doi: 10.1071/EN04076. [DOI] [Google Scholar]
- 45.Gu W, Ul Haque MF, Baral BS, Turpin EA, Bandow NL, Kremmer E, Flatley A, Zischka H, DiSpirito AA, Semrau JD. 2016. A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl Environ Microbiol 82:1917–1923. doi: 10.1128/AEM.03884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas SA, Rodby KE, Roth EW, Wu JS, Gaillard JF. 2018. Spectroscopic and microscopic evidence of biomediated HgS species formation from Hg(II)-cysteine complexes: implications for Hg(II) bioavailability. Environ Sci Technol 52:10030–10039. doi: 10.1021/acs.est.8b01305. [DOI] [PubMed] [Google Scholar]
- 47.Lu X, Johs A, Zhao L, Wang L, Pierce EM, Gu B. 2018. Nanomolar copper enhances mercury methylation by Desulfovibrio desulfuricans ND132. Environ Sci Technol Lett 5:372–376. doi: 10.1021/acs.estlett.8b00232. [DOI] [Google Scholar]
- 48.Chen HM, Johnston RC, Mann BF, Chu RK, Tolic N, Parks JM, Gu B. 2017. Identification of mercury and dissolved organic matter complexes using ultra-high resolution mass spectrometry. Environ Sci Technol Lett 4:59–65. doi: 10.1021/acs.estlett.6b00460. [DOI] [Google Scholar]
- 49.Liang X, Lu X, Zhao J, Liang L, Zeng E, Gu B. 2019. Stepwise reduction approach reveals mercury competitive binding and exchange reactions within natural organic matter and mixed organic ligands. Environ Sci Technol 53:10685−10694. doi: 10.1021/acs.est.9b02586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.