FIG 2.
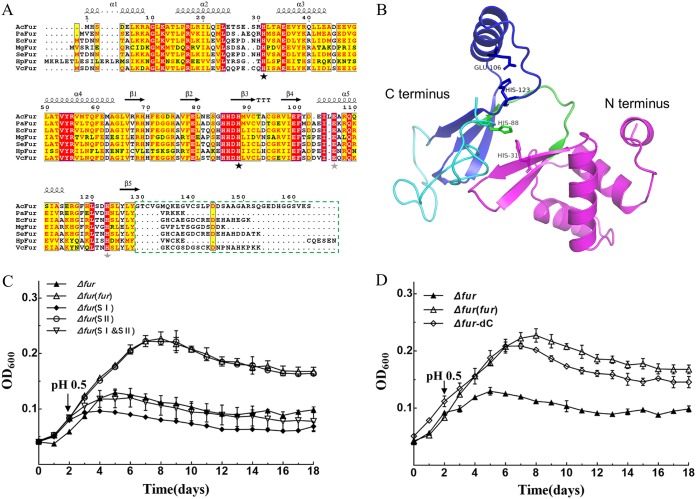
In silico analysis and functional confirmation of AcFur. (A) Multiple-sequence alignment of AcFur with Fur proteins from different bacteria by T-COFFEE and ESPript. The key residues of the metal-binding sites I and II are marked with black stars and gray stars under the sequences, respectively. The C-terminal extension of AcFur that was deleted to estimate its functionality is highlighted with a dashed box. (B) The AcFur model was generated by Phyre2 and visualized with PyMOL 2.0, as described in Materials and Methods. The model of the AcFur structure is shown in cartoon mode (N-terminal, magenta; C terminus, blue; linker, green; C-terminal extension, cyan). The putative key residues are shown as a stick model. (C and D) Phenotypic characterization of Δfur, Δfur(fur), Δfur(SI), Δfur(SII), Δfur(SI&SII), and Δfur-dC strains under acid shock. Error bars show standard deviations.