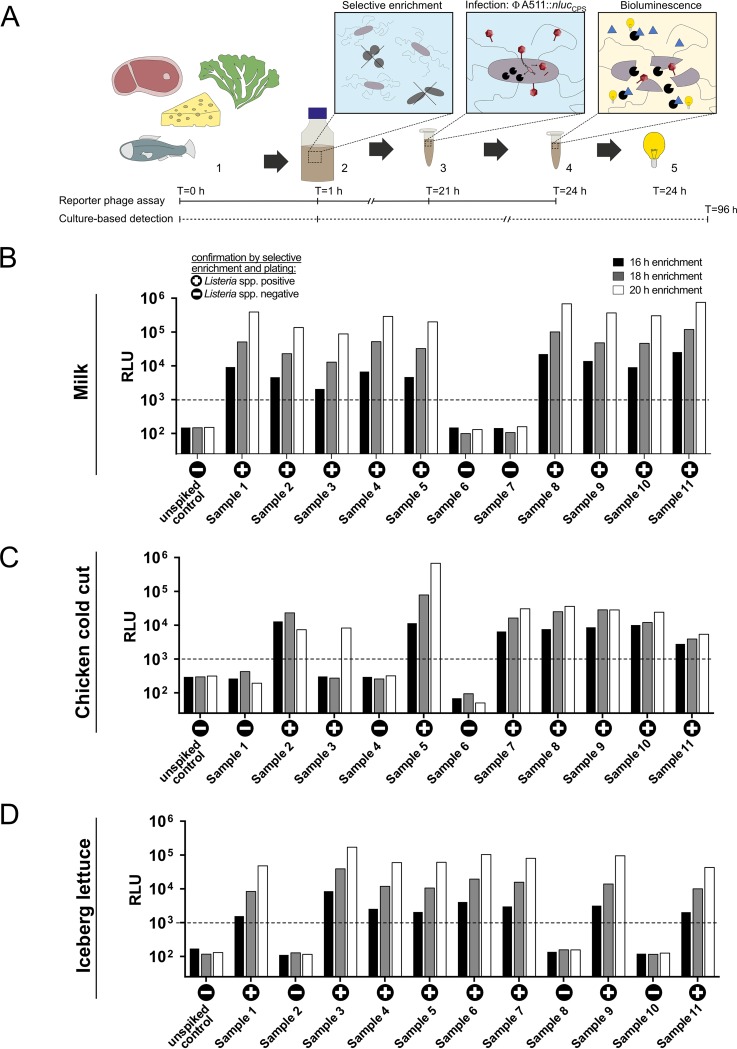
FIG 5.
Bioluminescence-based detection of Listeria in artificially contaminated foods. (A) Schematic representation of the workflow and duration of the reporter phage-based detection assay compared to classical culture-dependent detection. (1) Preparation of the food sample; (2) Listeria-selective enrichment; (3) phage infection; (4) substrate addition; (5) detection of bioluminescence. (B to D) Bioluminescence output (RLUs) of artificially spiked food samples (samples 1 to 11) and unspiked controls is shown after selective enrichment in 1/4 LEB (16, 18, and 20 h) and compared to results from a culture-based detection approach performed with the same samples: +, culture positive for Listeria; −, culture negative for Listeria. Samples were contaminated with ± 1 CFU/25 g of food. Quantified values are 1.2 ± 0.2 in milk, 1.1 ± 0.04 in chicken cold cut (errors are SEM of biological duplicates), and 1.0 ± 0.2 in iceberg lettuce (error is standard deviation of technical triplicates). Dotted lines indicate threshold value.