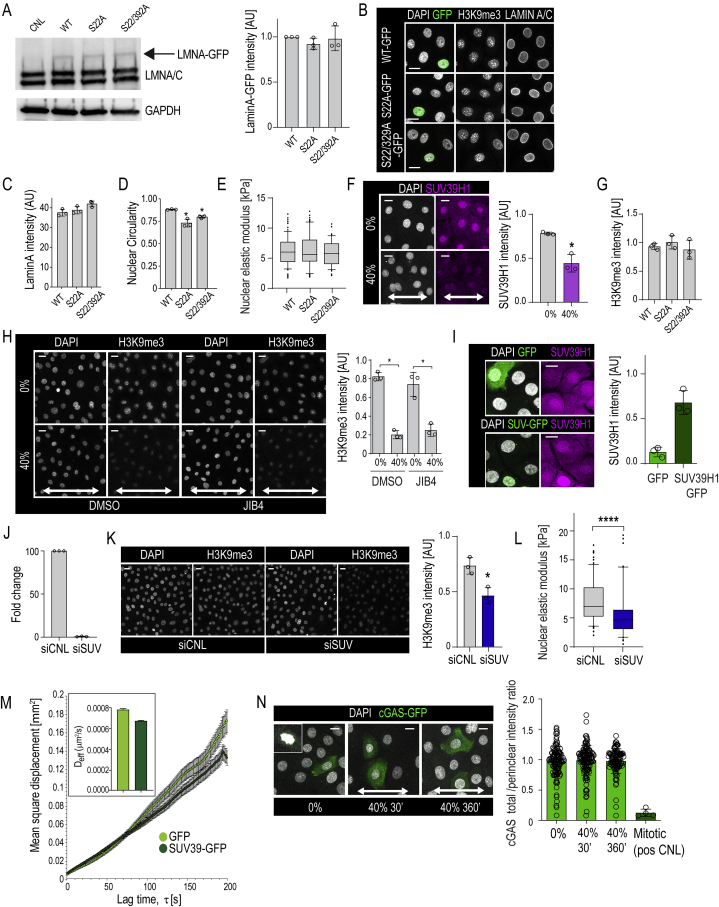
Figure S4.
H3K9me3 Regulates Chromatin Rheology and Nuclear Mechanics, Related to Figure 4
(A) Representative western blots and quantification of GFP-tagged wild-type (WT) Lamin A or Lamin A mutants (Serine 22 to Alanine (S22A) or S22A/S392A double mutant) show equal levels of expression. Lamin A band intensities are normalized to GAPDH (n = 3 independent experiments).
(B) Representative immunofluorescence images of nuclei (DAPI), GFP, Lamin A/C, and H3K9me3 in cells expressing WT Lamin A-GFP or Lamin mutants S22A-GFP; S22A/S392A-GFP.
(C and D) Quantification of Lamin A intensity and nuclear shape in WT Lamin A and mutant expressing cells (n = 3 independent experiments with > 100 nuclei/condition/experiment; ∗p = 0.034, ANOVA/Dunnett’s).
(E) AFM force indentation experiments of cells expressing WT Lamin A-GFP or Lamin mutants S22A-GFP; S22A/S392A-GFP (n = 3 independent experiments with > 40 nuclei/condition/experiment; no statistically significant difference found, Friedman/Dunn’s).
(F) Representative immunofluorescence images and quantification of Suv39H1 levels in cells exposed to uniaxial 40% stretch for 30 min (n = 3 independent experiments with > 100 nuclei/condition/experiment; ∗p = 0.05, Mann-Whitney)
(G) Quantification of H3K9me3 intensity from cells in (B) (n = 3 independent experiments with > 30 nuclei/condition/experiment; no statistically significant difference found, Friedman/Dunn’s).
(H) Representative immunofluorescence images and quantification of H3K9me3 levels in cells treated with JIB4 to inhibit H3K9me3 demethylation and exposed to 30 min uniaxial 40% stretch (n = 3 independent experiments with > 200 nuclei/condition/experiment; ∗p = 0342, Friedman/Dunn’s).
(I) Representative immunofluorescence images and quantification of Suv39H1 levels in cells transfected with Suv39H1-IRES-GFP or GFP only (n = 3 independent experiments with > 24 nuclei/condition/experiment).
(J) Quantitative RT-PCR analyses of Suv39H1 mRNA expression, normalized to B2M in EPC monolayers where Suv39H1 has been silenced by siRNA (siSUV) (n = 3 independent experiments).
(K) Representative immunofluorescence images and quantification of H3K9me3 in siSUV cells cells (n = 3 independent experiments with > 200 nuclei/condition/experiment; ∗p = 0.05, Mann-Whitney).
(L) AFM force indentation experiments of siSUV cells (n = 3 independent experiments with > 40 nuclei/condition/experiment; ∗∗∗∗p < 0.001, Mann-Whitney).
(M) Quantification of chromatin rheology by mean square displacement versus lag time τ of CRISPR-rainbow labeled telomeres. From linear data, material properties of chromatin can be calculated from MSD = Deffτ, where a higher Deff corresponds to less condensed chromatin (n = 40 (GFP)/30 (Suv39-GFP) cells from 3 independent experiments; ∗∗∗p = 0.0001, Student’s t test).
(N) Representative immunofluorescence images and quantification of cGAS-transfected cells to detect nuclear rupture in cells exposed to 30 or 360 min uniaxial 40% stretch. Note lack of perinuclear accumulation of cGAS indicating absence of nuclear rupture upon stretch. Inset shows accumulation of cGAS to chromatin upon mitotic breakdown of NE as positive control (n = 3 independent experiments with > 150 nuclei/condition/experiment). Bar graphs show mean ± SD, boxplots show 95% confidence interval, scale bars represent 10 μm, white arrows indicate stretch direction, AU = arbitrary units.