Figure 6.
Temporal Variability in Gene Expression
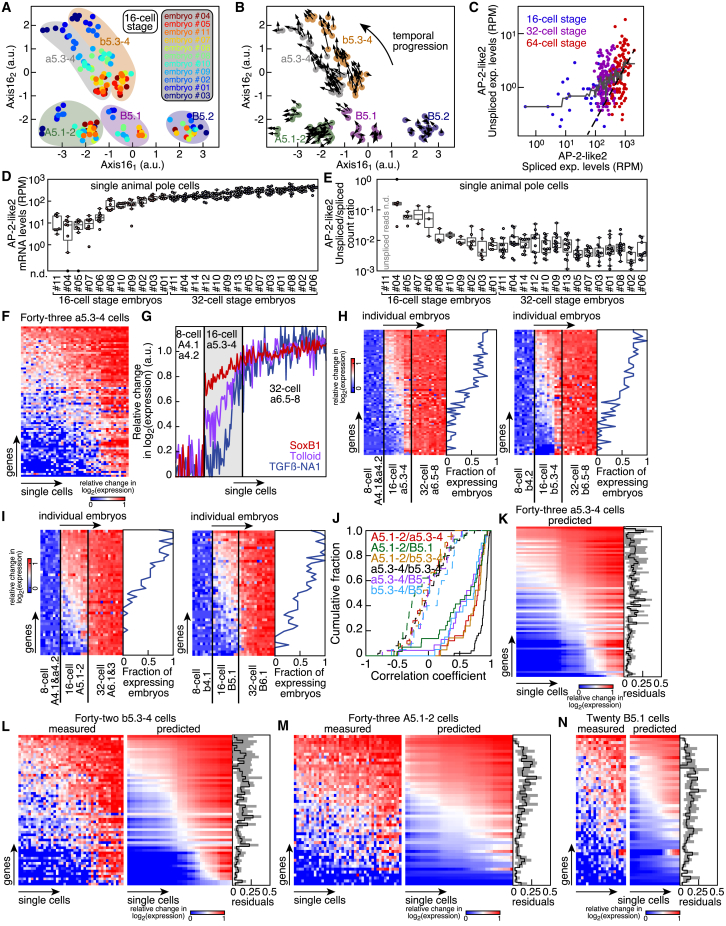
(A) Projection on Axis161 and Axis162 of single-cell transcriptomes of 11 16-cell embryos. Cell color according to embryo.
(B) RNA velocity field projected on Axis161 and Axis162. Arrows show the local velocity of individual cells (with measurable intron counts) of 16-cell embryos. Cell color according to cell type.
(C) Comparison between spliced and unspliced read counts of AP-2-like2 in single cells at the 16-, 32-, or 64-cell stage (blue, purple, and red respectively; gray, median filter; dashed black line, γ fit; STAR Methods).
(D and E) Expression levels (D) and ratio of unspliced to spliced read counts (E) of AP-2-like2 in single animal pole cells of individual embryos (n.d., not detected).
(F) Relative expression levels of upregulated genes in a5.3-4 cells.
(G) Relative expression levels of SoxB1, Tolloid, and TGFβ-NA1 in a5.3-4 cells, their mother and daughter cell types.
(H and I) Relative expression levels of genes upregulated in the animal (H) and vegetal somatic cell types (I) (excluding the germ cell lineage) of 16-cell stage embryos, their mother and daughter cell types. Levels were averaged in individual embryos. The same order of embryos was used for the 4 heatmaps. The fraction of 16-cell embryos expressing each gene at a level greater than 0.5 × the log-transformed average expression level at the 32-cell stage is indicated.
(J) Correlation coefficient of expression levels of genes that were upregulated in all the combination of two somatic cell types at 16-cell stage (solid lines; dotted lines, randomized embryos).
(K) Relative expression levels of upregulated genes in a5.3-4 cells predicted with a model of linear variation in time (STAR Methods). Residuals for individual genes are indicated (black line, median; gray shade, interquartile range).
(L–N) Relative expression levels of upregulated genes as measured by scRNA-seq or predicted with a model of linear variation in time in b5.3-4 (L), A5.1-2 (M), and B5.1 (N) cells. Residuals for individual genes are indicated (black line, median; gray shade, interquartile range).
See also Figure S7.