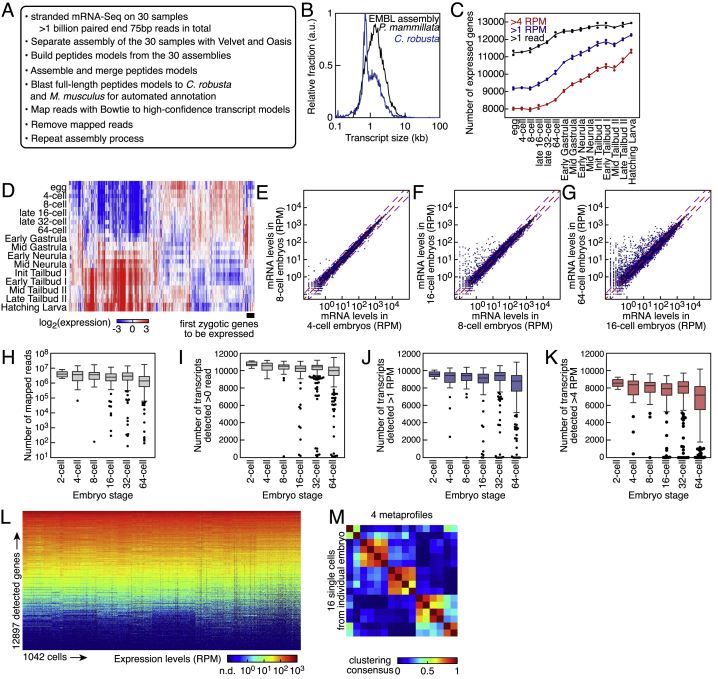
Figure S1.
P. mammillata Developmental Transcriptome, Related to Figure 1
(A) Workflow for the de novo assembly of P. mammillata transcriptome. Ciona robusta was formerly known as Ciona intestinalis.
(B) Distribution of P. mammillata transcript lengths. As comparison, the distribution for C. robusta (formerly known as C. intestinalis) is shown in blue.
(C) Number of detected genes during development. Threshold in normalized reads (RPM: reads per million mapped reads): 1 (blue) and 4 (red) RPM. Black line: threshold at one mapped read. Staging was performed according to Hotta et al. (2007).
(D) Hierarchical clustering of mRNA expression during P. mammillata embryogenesis. Staging was performed according to Hotta et al. (2007).
(E) Expression profiles in 4- and 8-cell stage embryos as measured by RNA-Seq. Pink dashed lines indicate 2-fold expression changes.
(F) Expression profiles in 4- and 16-cell stage embryos as measured by RNA-Seq. Pink dashed lines indicate 2-fold expression changes.
(G) Expression profiles in 16- and 64-cell stage embryos as measured by RNA-Seq. Pink dashed lines indicate 2-fold expression changes.
(H) Number of mapped reads for the 1084 single cells pooled by developmental stage.
(I) Number of transcripts detected with at least one read across the 1042 single-cell transcriptomes pooled by developmental stage (RPM: reads per million mapped reads).
(J) Number of transcripts with expression exceeding 1 read per million (RPM) across the 1042 single-cell transcriptomes pooled by developmental stage.
(K) Number of transcripts with expression exceeding 4 reads per million (RPM) across the 1042 single-cell transcriptomes pooled by developmental stage.
(L) Expression levels of 12,897 detected genes across 1042 single cells. Genes were sorted according to the median expression level across all cells.
(M) Non-negative matrix factorization of gene expression profiles of the 16 single cells forming an individual 16-cell stage embryo. 4 metaprofiles split the embryo into the same 5 groups of 4, 4, 2, 4 and 2 cells with degraded consensus clustering compared to three metaprofiles.