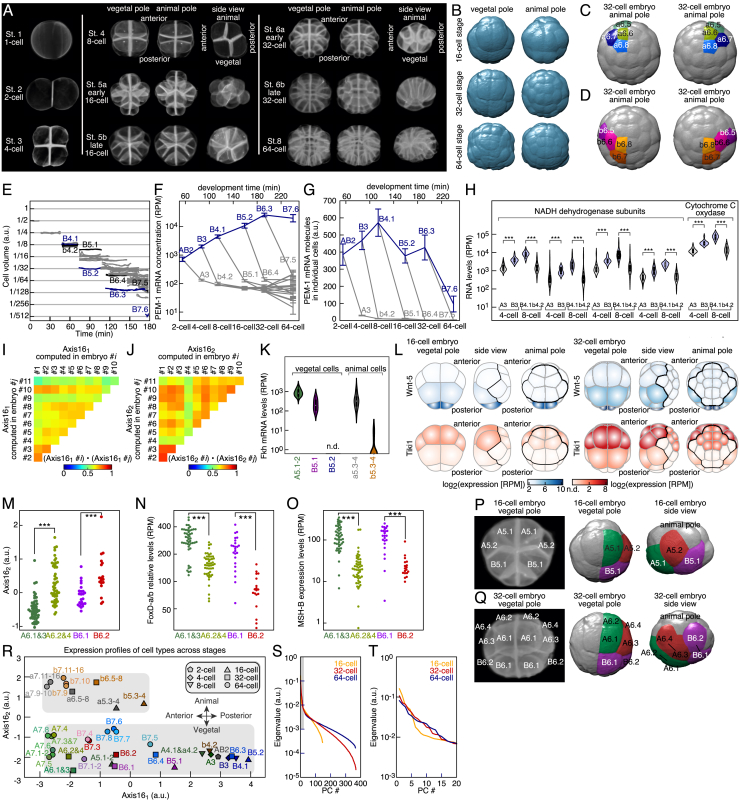
Figure S4.
Early Development of P. mammillata, Asymmetric RNA Inheritance and Spatial Mapping of scRNA-Seq Data, Related to Figure 4
(A) Live imaging of P. mammillata early development. Images of embryos expressing membrane-bound Citrine were rendered using Chimera (Pettersen et al., 2004).
(B) Morphology of P. mammillata embryos. Segmented membrane signal of the vegetal and animal poles at 16-, 32 and 64-cell stages.
(C) Progeny of the two a4.2 cells (one for each side of the embryo) after two rounds of division at the 32-cell stage forming two pairs of sister cells: a6.5 and a6.6 (green) and a6.7 and a6.8 (blue).
(D) Progeny of the two b4.2 cells (one for each side of the embryo) after two rounds of division at the 32-cell stage forming two pairs of sister cells: b6.5 and b6.6 (purple) and b6.7 and b6.8 (orange/red).
(E) Temporal evolution of the cellular volumes as measured by light sheet microscopy. Blue: germ cell lineage (B4.1, B5.2, B6.3, B7.6); black: corresponding sister cells (b4.2, B5.1, B6.4, B7.5 respectively; gray: other cells.
(F) Expression levels of the maternal factor PEM-1 as measured by scRNA-Seq. Blue line: germ cell lineage (giving rise to cell B7.6 at 64-cell stage). Gray lines: all the other lineages. Data represented as mean ± SEM.
(G) Total mRNA levels of the maternal factor PEM-1 as measured by scRNA-Seq and scaled by cellular volume measured by light sheet microscopy. Blue line: germ cell lineage (giving rise to cell B7.6 at 64-cell stage). Gray lines: all the other lineages. Data represented as mean ± SEM.
(H) Violin plots of expression levels for five mitochondrial transcripts coding for four subunits of the NADH dehydrogenase and cytochrome C oxydase. ∗∗∗: p < 0.001, Kruskal-Wallis test.
(I, J) Comparison of the eleven different Axis161 (I) and Axis162 (J) determined independently for the eleven 16-cell embryos assessed. p < 10−32 for Axis161 and p < 10−46 for Axis162 to get the reported degree of colinearity between two random vectors in the 595 dimensions used to compute the principal component analysis.
(K) Violin plot of the forkhead transcription factor Fkh expression levels at 16-cell stage (n.d.: not detected).
(L) Expression levels of Wnt-5 (blue) and the metalloprotease Tiki1 (red) at the 16-cell stage and the 32-cell stage as measured by scRNA-Seq (n.d: not detected).
(M) Projection on Axis162 of single-cell transcriptomes of 47 A6.1&3, 48 A6.2&4, 24 B6.1 and 18 B6.2 cells (Comparison between A6.1&3 and A6.2&4 cells: p = 5 × 10−11; Comparison between B6.1 and B6.2 cells: p = 10−4, Kruskal-Wallis test).
(N) FoxD-a/b expression levels in 47 A6.1&3, 48 A6.2&4, 24 B6.1 and 18 B6.2 cells (Comparison between A6.1&3 and A6.2&4 single cells: p = 7 × 10−11; Comparison between B6.1 and B6.2 cells: p = 10−6, Kruskal-Wallis test).
(O) MSH-B expression levels in 47 A6.17&3, 48 A6.2&4, 24 B6.1 and 18 B6.2 cells (Comparison between A6.1&3 and A6.2&4 single cells: p = 5 × 10−13; Comparison between B6.1 and B6.2 cells: p = 4 × 10−6, Kruskal-Wallis test).
(P, Q) Membrane signal and segmented membrane signal of the vegetal pole at 16- (P) and 32-cell stages (Q). A6.2, A6.4 and B6.2 lie closer to the animal pole compared to their respective sister cells A6.1, A6.3 and B6.1. Sister cells are linked by bars and have the same color as their mother.
(R) Projection on Axis161 and Axis162 of average expression profiles for all cell types identified.
(S, T) Eigenvalue spectrum of the principal components (PC) computed with scRNA-Seq expression profiles of 16-, 32- or 64-cell embryos (yellow, red and blue respectively). Full spectrum (S) or zoom on the first 20 PCs (T, corresponding to the gray shaded area in S) are shown.