Figure 7.
A GATA Switch May Underlie Changes in Gene Expression in the Development of Splenic Red Pulp Macrophages (RPMs)
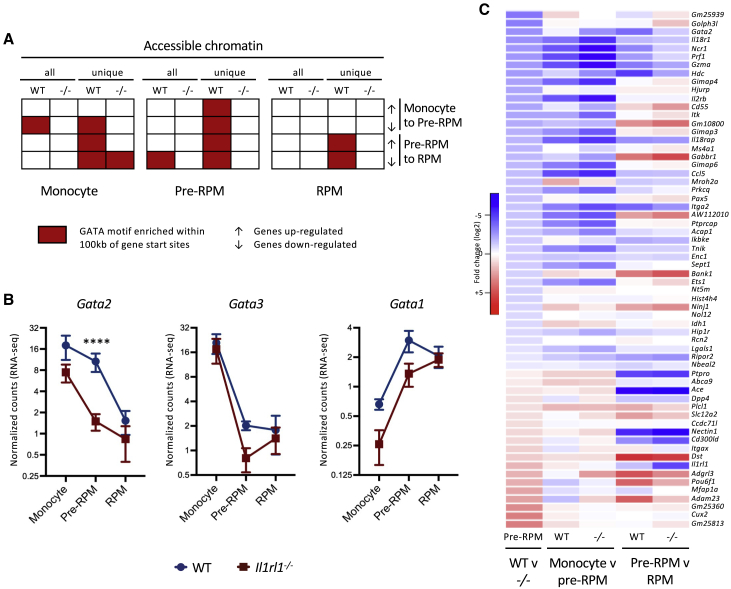
(A) Identification of GATA motifs in accessible chromatin of splenic monocytes (CD11bhi F4/80−), pre-RPMs (CD11bhi F4/80lo), and RPMs (CD11blo/− F4/80hi), within 100 kb of the start sites of genes differentially expressed between WT monocytes and WT pre-RPMs, or WT pre-RPMs and WT RPMs (both log2 fold change > 1, adjusted p value < 0.05). Motif enrichment was performed on accessible chromatin from each splenocyte population from WT and Il1rl1−/− mice and also from accessible chromatin from each splenocyte population that was unique to WT or unique to Il1rl1−/− mice; n = 4 separate mice per group.
(B) Relative Gata gene expression (normalized counts, cpm) in splenic monocytes (CD11bhi F4/80−), pre-RPMs (CD11bhi F4/80lo), and RPMs (CD11blo/− F4/80hi) of WT (green) and Il1rl1−/− mice (red); n = 4 separate mice per group. Data are represented as mean ± SEM. ∗∗∗∗ adjusted p (WT v −/−) < 0.001.
(C) Heatmaps showing log2 fold changes of genes differentially expressed in WT and Il1rl1−/− pre-RPMs (adjusted p value < 0.05) between splenic monocytes (CD11bhi F4/80−) and pre-RPMs (CD11bhi F4/80lo), or pre-RPMs (CD11bhi F4/80lo) and RPMs (CD11blo/− F4/80hi), from WT and Il1rl1−/− mice; n = 4 per group.
Please see also Figure S7.