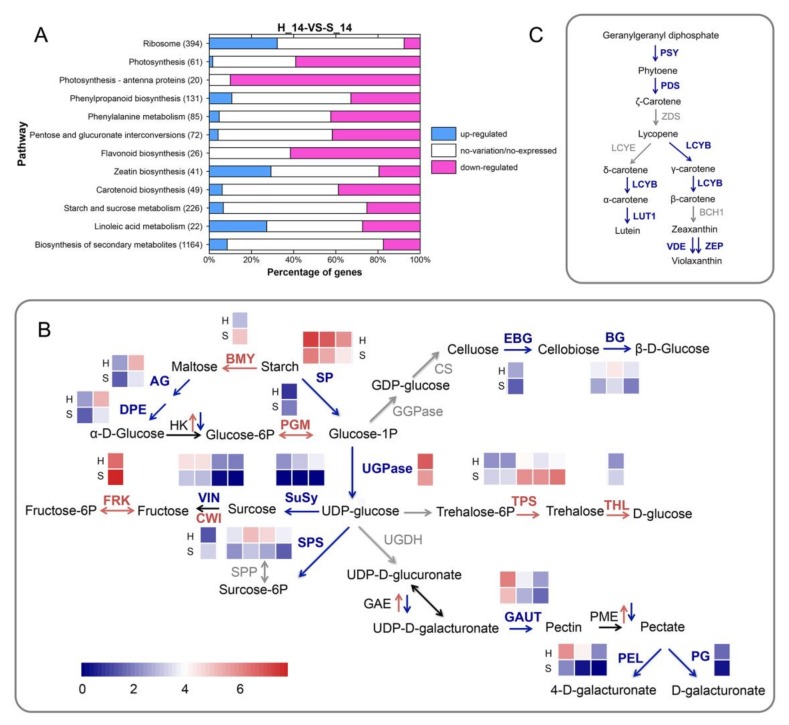
Figure 6.
Differentially expressed genes (DEGs) involved in biological pathways. (A) The top 12 metabolic pathways with significant enrichment of DEGs. The numbers in parentheses represent the numbers of DEGs with pathway annotations. (B) Illustration of the enzymes encoded by DEGs involved in carbohydrate metabolism. Note: AG, alpha-glucosidase (EC 3.2.1.20); BG, β-glucosidase (EC 3.2.1.21); BMY, β-amylase (EC 3.2.1.2); CWI, cell wall invertase; CS, cellulose synthase (guanosine diphosphate (GDP)-forming) (EC 2.4.1.29); DPE, 4-α-glucanotransferase (EC 2.4.1.25); EBG, endo-1, 4-beta-glucanase (EC 3.2.1.4); FRK, fructokinase (EC 2.7.1.4); GAE, UDP-glucuronate 4-epimerase (EC 5.1.3.6); GAUT, galacturonosyltransferase (EC 2.4.1.43); GGPase, GDP glucose pyrophosphorylase (EC 2.7.7.34); HK, hexokinase (EC 2.7.1.1); PEL, pectate lyase (EC 4.2.2.2); PG, polygalacturonase (EC 3.2.1.15); PGI, glucose-6-phosphate isomerase (EC 5.3.1.9); PGM, phosphoglucomutase (EC 5.4.2.2); SP, starch phosphorylase (EC 2.4.1.1); SPP, sucrose phosphate phosphatase (EC 3.1.3.24); SuSy, sucrose synthase (EC 2.4.1.13); PME, pectin methyl esterase (EC 3.1.1.11); THL, trehalase (EC 3.2.1.28); TPS, α, α-trehalose-phosphate synthase (uridine diphosphate (UDP)-forming) (EC 3.1.3.12); UGDH, UDP-glucose 6-dehydrogenase (EC 1.1.1.22); UGPase, UDP-glucose pyrophosphorylase (EC 2.7.7.9); VIN, soluble acid invertase (EC 3.2.1.26). Red and blue fonts represent up- and downregulation of gene expression, respectively. Each box depicts an individual gene. Enzymes encoded by upregulated, downregulated, and non-differentially expressed genes are shown in red, blue, and gray, respectively. The scale bar represents reads per kilobase of exon model per million mapped reads (RPKM) values. (C) DEGs involved in carotenoid biosynthesis. Note: BCH1, carotenoid β-hydroxylase (EC 1.14.15.24); LCYB, lycopene β-cyclase (EC 5.5.1.19); LCYE, lycopene ε-cyclase (EC 5.5.1.18); LUT1, cytochrome P450-type monooxygenase (EC 1.14.14.158); PDS, phytoene desaturase (EC 1.3.5.5); PSY, phytoene synthase (EC 2.5.1.32); VDE, violaxanthin de-epoxidase (EC 1.10.99.3); ZDS, ζ-carotene desaturase (EC 1.14.99.30); ZEP, zeaxanthin epoxidase (EC 1.14.13.90). Enzymes encoded by non-differentially expressed and downregulated genes are shown in gray and blue, respectively.