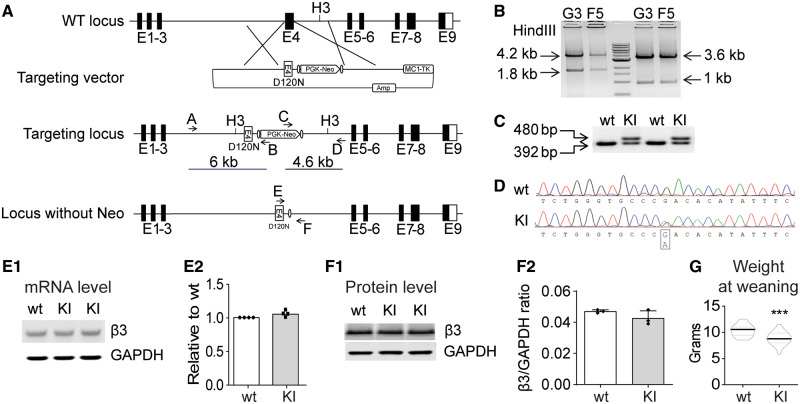
Figure 1.
Creation of the Gabrb3+/D120N KI mouse. (A) Scheme for targeted KI of the Gabrb3 locus. A vector was constructed to replace Gabrb3 Exon 4 genomic DNA with an Exon 4 containing the G358A mutation and the positive selection cassette (PGK-Neo). (B) Long-range PCR was used to identify the correctly targeted clones, G3 and F5. The 6- and 4.6-kb fragments were amplified from correctly targeted clones using primers A/B and C/D, respectively. The PCR fragments were digested with the HindIII restriction enzyme (to generate the predicted 4.2- and 1.8-kb fragments for the 5′ end and 3.6- and 1-kb fragments for the 3′ end) to further confirm correct homologous recombination. (C) The KI mice were genotyped. PCR amplified a 392-bp fragment from the wild type β3 subunit allele and a 480-bp fragment from the β3 subunit KI allele due to the insertion of FRT sequences and other modifications. (D) Sequencing chromatogram of RT-PCR derived from total brain RNA showed the presence of the G358A mutation and corresponding equal level of wild type and mutant nucleotide expression. (E1) Semi-quantitative RT-PCR showed that the KI allele did not affect Gabrb3 gene expression, using GAPDH as a loading control. (E2) Band intensities of the RT-PCR products were first normalized to GAPDH for quantification and then to wild type levels. (F1) Immunoblotting showed equal β3 subunit expression in KI and wild type littermate mouse brain whole-cell lysates. GAPDH served as a loading control. (F2) Protein levels were normalized first to internal GAPDH and then to wild type levels. (G) The body weight of KI and their littermate mice at weaning were plotted (n = 22 wild type, 18 KI). GAPDH: glyceraldehyde 3-phosphate dehydrogenase; RT-PCR: reverse transcriptase PCR.