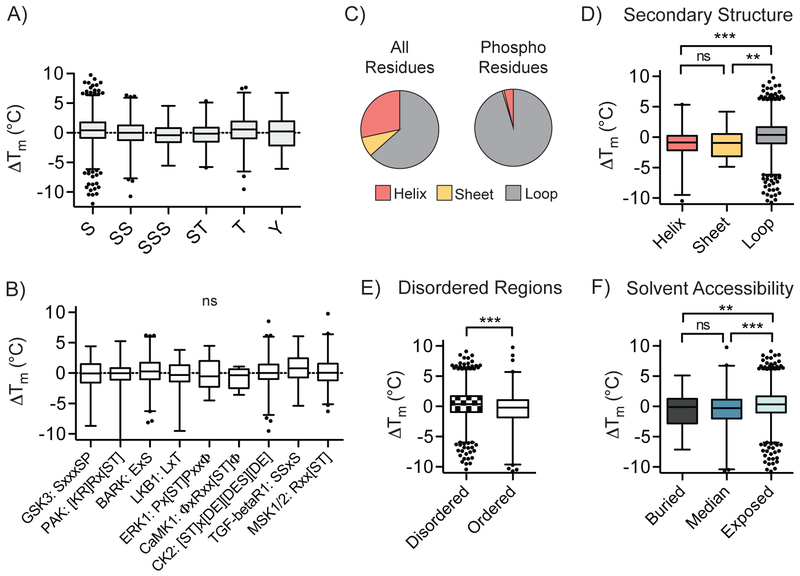
Fig. 3. Global relationships between local phosphosite environment and altered phosphomodiform stability.
A) Distribution of ΔTm values for tryptic peptides containing indicated phosphoamino acids or coincidental combinations thereof (P < 0.0001, one-way ANOVA). B) Correlation between detected kinase substrate motifs and ΔTm values for identified sites (P = 0.3011, one-way ANOVA). C) Distribution of all amino acids in the human proteome (left) and detected phosphosites (right) in predicted secondary structural elements. D-F) Comparisons of ΔTm values and predicted secondary structure elements (D; ***, P ≤ 0.0001, two-sided t-test), ordered structural elements surrounding the phosphosite of interest (E; ***, P ≤ 0.0001, two-sided t-test), and solvent accessibility (F; ***, P ≤ 0.0001, two-sided t-test). Box plots (median, 1-99%) are shown in (A-B, D-F), with outliers shown as data points. All data were generated from n = 12 (bulk, unmodified) and n = 10 (phospho) total MS technical replicates from n = 6 (bulk, unmodified) and n = 5 (phospho) independent biological replicates.