Figure 4: The SARS-CoV-2 interactome reveals novel aspects of SARS-CoV-2 biology that can be targeted pharmacologically.
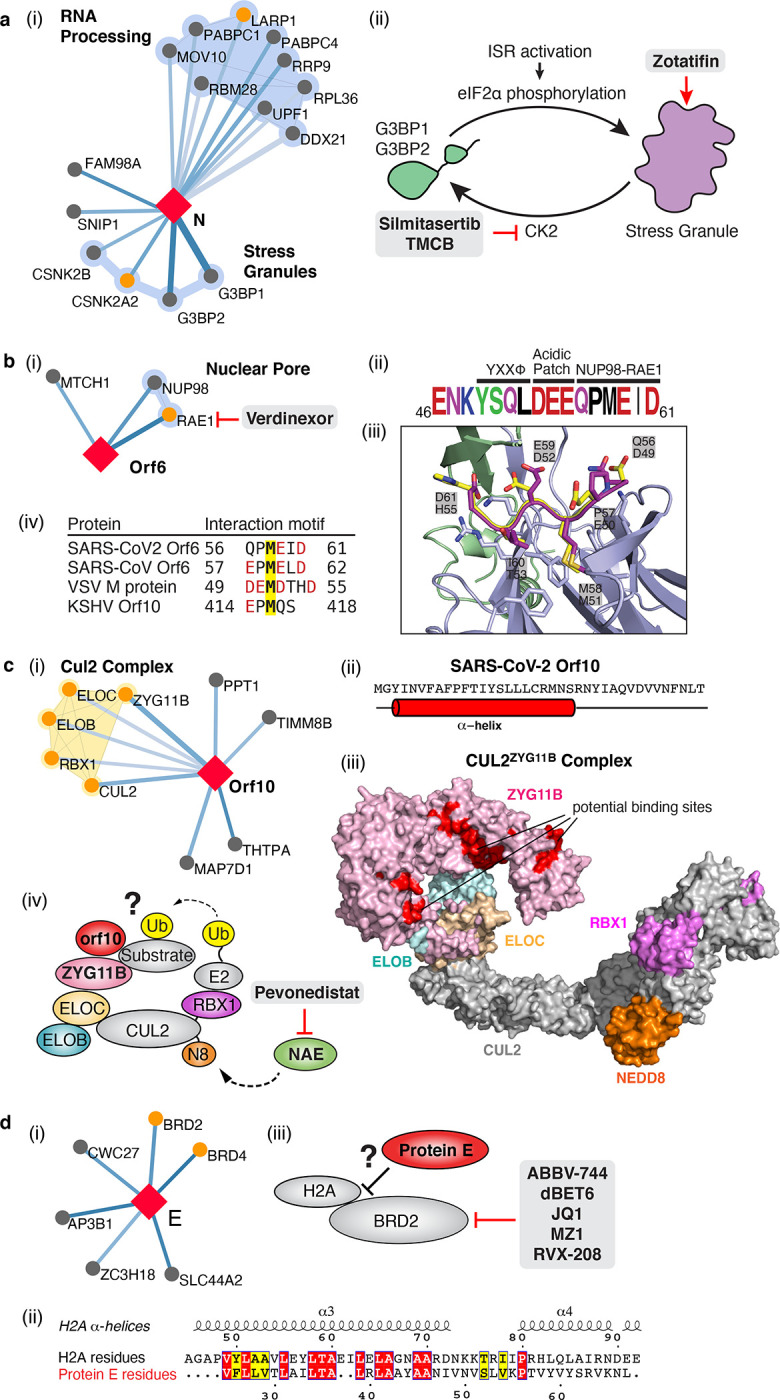
(a) Protein N targets stress granule proteins. (i) Protein N interactome. (ii) Model for therapeutic targeting of N interactions in the formation of stress granules (SGs). SGs are known to exhibit antiviral activity, with the integrative stress response (ISR) inducing eIF2α phosphorylation and SG formation, and Casein kinase II (CK2) disrupting and preventing the formation of SGs. By activating SG formation, or inhibiting CK2, the cellular environment could potentially shift to a more antiviral state. (b) Orf6 interacts with an interferon-inducible mRNA nuclear export complex. (i) Orf6 interactome including small molecule inhibitors for RAE. (ii) Annotated C-terminal sequence of SARS-CoV-2 Orf6, highlighting previously described trafficking motifs and the putative NUP98-RAE1 binding sequence. Colors indicate chemical properties of amino acids: polar (G,S,T,Y,C, green), neutral (Q,N, purple), basic (K, R, H, blue), acidic (D, E, red), and hydrophobic (A, V, L, I, P, W, F, M, black). (iii) SARS-CoV-2 Orf6 carboxy-terminal peptide modeled into the binding site of the VSV M protein-NUP98-RAE1 complex (PDB ID: 4OWR). Orf6 shown in dark purple, M protein in yellow, NUP98 in green, and RAE1 in light purple. Orf6 and M protein residues labeled. RAE1 hydrophobic residues contacting the key methionine and basic patch residues of RAE1 and NUP98 are shown. (iv) Putative NUP98-RAE1 interaction motifs present in proteins from several viral species. The consensus motif consists of negatively charged residues (red) surrounding a conserved methionine (yellow). (c) Orf10 interacts with the CUL2ZYG11B complex. (i) Orf10 interactome. (ii) The secondary structure of Orf10 contains an alpha helix motif. (iii) Surface representation of the homology model for CUL2ZYG11B complex, residues that are conserved amongst ZYG11B orthologues from various species are indicated in red are likely protein interaction surfaces for binding substrates and other proteins. (iv) A possible model of how Orf10 binds to the CUL2ZYG11B complex to hijack the complex for ubiquitination or viral restriction factors and how it can be targeted pharmacologically. (d) Envelope (E) interacts with bromodomain proteins. (i) E interactome. (ii) Sequence alignment of highlighted regions of E and Histone 2A (H2A). The positions with identical and similar amino acid residues are highlighted in red and yellow, respectively. Note the greater hydrophobicity of E may indicate a part of the alignment represents a transmembrane segment. (iii) Model of how E might mimic the BRD2 native interaction partner Histone 2A and how BRD2 can be targeted pharmacologically.
