Figure 2. Detection of SARS-CoV-2 in contrived and clinical nasopharyngeal or oropharyngeal swab samples.
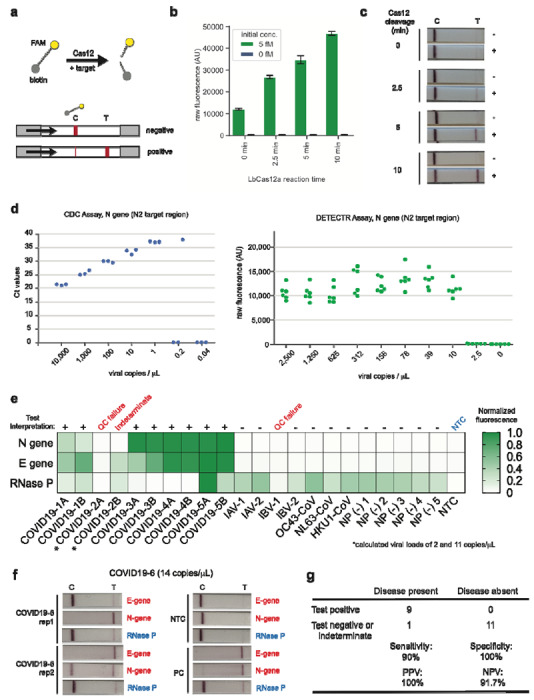
(a) Schematic of DETECTR coupled with lateral flow readout. The intact FAM-biotinylated reporter molecule flows to the control capture line. Upon recognition of the matching target, the Cas-gRNA complex cleaves the reporter molecule, which flows to the target capture line. (b-c) Comparison of fluorescence to lateral flow. (b) Fluorescence signal of LbCas12a detection assay on RT-LAMP amplicon for SARS-CoV-2 N-gene saturates within 10 min. RT-LAMP amplicon generated from 2 μL of 5 fM or 0 fM SARS-CoV-2 N-gene IVT RNA by amplifying at 62°C for 20 min. (c) LbCas12a on the same RT-LAMP amplicon produces visible signal through lateral flow assay within 5 min. (d) Limit of detection for CDC qPCR and DETECTR. Ct values using the CDC qPCR assay (n=3) and fluorescence values using SARS-CoV-2 DETECTR (n=6) using SARS-CoV-2 N2 gene IVT-RNA. (e) Patient sample DETECTR data. DETECTR fluorescence values were normalized to the highest value within the N gene, E gene or RNase P set, with a positive threshold at five standard deviations above background. Final determination of the SARS-CoV-2 test was based on the interpretation matrix in Fig. 1e, with results indicated above the heat map. (f) SARS-CoV-2 DETECTR assay identifies presence of SARS-CoV-2 viral RNA from clinical sample. Two replicate assays were performed using 2 μL of extracted RNA for each reaction (titer 12 copies/μL). Positive controls used IVT RNA for SARS-CoV-2 targets and total human RNA for RNase P. LbCas12a detection assays were run on lateral flow strips (TwistDx) and imaged after 3 min. (g) Performance characteristics of the SARS-CoV-2 DETECTR assay. Abbreviations: fM, femtomolar; NTC, no-template control; PPV, positive predictive value; NPV, negative predictive value.
