Abstract
Cadmium (Cd) is a toxic metal element and the mechanism(s) underlying Cd tolerance in plants are still unclear. Increasingly more studies have been conducted on Cd binding to plant cell walls (CW) but most of them have focused on Cd fixation by CW pectin, and few studies have examined Cd binding to cellulose and hemicellulose. Here we found that Cd binding to CW pectin, cellulose, and hemicellulose was significantly higher in Tor-1, a Cd tolerant A. thaliana ecotype, than in Ph2-23, a sensitive ecotype, as were the concentrations of pectin, cellulose, and hemicellulose. Transcriptome analysis revealed that the genes regulating CW pectin, cellulose, and hemicellulose polysaccharide concentrations in Tor-1 differed significantly from those in Ph2-23. The expressions of most genes such as pectin methyl esterase inhibitors (PMEIs), pectin lyases, xyloglucan endotransglucosylase/hydrolase, expansins (EXPAs), and cellulose hydrolase were higher in Ph2-23, while the expressions of cellulose synthase-like glycosyltransferase 3 (CSLG3) and pectin ethyl esterase 4 (PAE4) were higher in Tor-1. The candidate genes identified here seem to regulate CW Cd fixation by polysaccharides. In conclusion, an increase in pectin demethylation activity, the higher concentration of cellulose and hemicellulose, regulated by related genes, in Tor-1 than in Ph2-23 are likely involved in enhanced Cd CW retention and reduce Cd toxicity.
Keywords: cadmium tolerance, Arabidopsis thaliana ecotypes, phytoremediation, cell wall polysaccharides, pectin, cellulose, hemicellulose
Introduction
Cadmium (Cd) is a toxic, heavy metal element that hinders plant growth and causes potential health risks to humans through the food chain (Hu et al., 2013; Åkesson et al., 2014). It is listed among the top 10 most dangerous substances by the Agency for Toxic Substances and Disease Registry (Benton et al., 2011). When excess Cd enters the cytoplasm of plant cells, it causes toxicity by triggering many oxidative stress reactions, such as lipid peroxidation, protein denaturation (Rellán-Álvarez et al., 2006), and increased reactive-oxygen species (Jia et al., 2016), which affect plant growth. Therefore, it is important to thoroughly understand the tolerance mechanisms of Cd in plants.
Cadmium accumulation in plant shoots mainly depends on Cd uptake by roots, sequestration within root vacuoles, and transportation and redistribution from roots to shoots (Ismael et al., 2019). Plant tolerance to Cd is based on various mechanisms, such as Cd chelation by cell wall (CW) components, vacuole compartmentalization, and Cd chelation by cytoplasmic organic acids or peptides (Memon and Schröder, 2009; Zhang J. et al., 2018; Deng et al., 2019).
The CW is a major storage site for heavy metals (Lux et al., 2013). It acts as the first barrier against Cd stress, as well as a protective barrier of protoplasts, mainly by directly trapping Cd outside the cell, which reduces cellular damage due to Cd toxicity (Richter et al., 2017). The primary CW is mainly composed of cellulose, hemicellulose, and pectin; some functional proteins, and a small amount of aromatic compounds (Showalter, 1993; Chebli and Geitmann, 2017). Among them, pectin polysaccharides modified by pectin methyl esterases (PMEs) and pectin acetyl esterases (PAEs) represent the most important CW components capable of binding to Cd (Peng et al., 2017), and pectin methyl esterase inhibitors (PMEIs) were reported to inhibit PMEs activity (Sénéchal et al., 2015). However, other polysaccharides in CWs also play important roles in metal binding and accumulation, such as cellulose and hemicellulose (Nishizono et al., 1987; Wan and Zhang, 2012). Data from previous studies showed that pectin polysaccharide levels in plant CWs increased under Cd stress (Meyer et al., 2015). Exogenous nitric oxide can enhance Cd tolerance in rice plants by increasing the pectin and hemicellulose contents in their root CWs (Xiong et al., 2009). In monocotyledonous and dicotyledonous non-grass plants, hemicellulose is mainly composed of xyloglucan. The xyloglucan endotransglucosylase/hydrolase enzyme (encoded by the XTH gene) catalyzes the hydrolysis and transglycosylation of xyloglucan polymers in plant CWs (Maldonado-Mendoza et al., 2005). Previous findings have shown that the XTH gene family may play important roles in alleviating metal toxicity in plants (Zhu et al., 2012). Loss of XTH31 function resulted in enhanced aluminum tolerance (Zhu et al., 2014) and the xth15 transfer-DNA mutant was more resistant to aluminum than the wild type (Zhu et al., 2013a). Although little research has described cellulose and Cd binding, a previous study showed that the Cd content in plant cellulose increased significantly after Cd treatment (Wang P. et al., 2018).
Recent studies on the CW retention of Cd have mainly focused on pectin, and few studies have been conducted to investigate hemicellulose and cellulose binding to Cd (Parrotta et al., 2015). In the present study, we found that genes such as PAEs, pectin lyases, XTHs, and cellulose synthase-like glycosyltransferase 3 (CSLG3) families participate in the regulation of pectin modification, as well as cellulose and hemicellulose polysaccharide levels. Based on transcriptome analysis, we also found these candidate genes may participate in regulating Cd chelation to the CW by altering the Cd concentration in different cellular components. As so, Cd could accumulate more in plant CWs, thereby reducing Cd toxicity to cellular organelles and providing a theoretical basis for Cd phytoremediation.
Materials and Methods
Plant Materials
Arabidopsis thaliana ecotypes Ph2-23 and Tor-1 were provided by Dr. Chao Daiying, Institute of Plant Physiology and Ecology, Shanghai Academy of Life Sciences, Chinese Academy of Sciences.
Growth Conditions
The two Arabidopsis thaliana ecotypes were seeded in a vegetative soil culture pot in a greenhouse (300 mmol photons m-2s-2, 16-h photoperiod, 22°C), sealed with a membrane to retain water, and the seeds germinated 3 days later. When the A. thaliana plants had grown two true leaves, the seedlings were transferred to a 4-L pot for hydroponic culture. The nutrient solution contained 1.25 mM KNO3, 0.625 mM KH2PO4, 0.5 mM MgSO4, 0.5 mM Ca(NO3)2⋅4H2O, 0.025 mM Fe-EDTA, 0.25 ml L–1 micronutrients (70 mM H3BO3, 14 mM MnCl2, 1 mM ZnSO4, 0.5 mM CuSO4, and 0.2 mM NaMoO4). The nutrient solution was renewed every 4 days. After 20 days of hydroponic growth, plants in the experimental group were treated with 10 μM CdCl2 for 4 days, and those in the control group were not treated with Cd.
Determination of Chlorophyll, Proline, and Malondialdehyde (MDA) Concentrations
Twenty-day-old A. thaliana plants were treated with or without 10 μM CdCl2 for 4 days and their rosette leaves (approximately 0.5 g) were extracted with 80% acetone and left in the dark for 24 h (Wang T. et al., 2018). The extracts were used for measuring absorption at 645 and 663 nm with a spectrophotometer. Chlorophyll a, chlorophyll b, and total chlorophyll concentrations were calculated as described by Arnon (1949). Proline concentrations were measured using ninhydrin colorimetry as described by Bates et al. (1973). Briefly, shoot tissues (0.5 g) were sampled, ground in 5 mL of 3% sulfosalicylic acid, and centrifuged at 22,000 × g for 5 min. The supernatant of each sample was collected, and proline concentration was determined after reaction with acid indene. The MDA concentration was determined by the thiobarbituric acid method (Esterbauer et al., 1991; Wang T. et al., 2018). Briefly, after full grinding in 5% trichloroacetic acid, the supernatant was centrifuged for 10 min at 3000 × g. Then, 2 mL of the supernatant was thoroughly mixed with 2 mL 0.67% thiobarbituric acid. The sample was then bathed at 100°C for 30 min. The absorbance of each supernatant was measured at 450, 532, and 600 nm after centrifugation.
Determination of Dry Weight, Total Cd Content, and Metal Concentrations Under Low Temperature
Twenty-day-old A. thaliana plants were treated with or without 10 μM CdCl2 for 4 days, after which their roots and shoots were harvested and washed with 0.1 mM CaCl2 for 1 min, followed by four times rinsing with deionized water, as described by Jian et al. (2019), and then dried in an oven to a constant mass (dry weight). For the total Cd assay, 20-day-old A. thaliana plants were transferred to Cd-free or 10 μM CdCl2 solutions for 4 days. After this period, the whole plants were collected, rinsed with deionized water four times and dried to constant weight. Then, 1 mL nitric acid was added, and both samples were digested in a boiling water bath for 2 h. Each sample was diluted 100-fold with deionized water (Jian et al., 2019), and the total Cd concentration and other metal concentrations were determined by inductively coupled plasma mass spectrometry (ICP-MS) on a NexIONTM 350X instrument (PerkinElmer, Massachusetts, United States). Total Cd content was calculated as total Cd concentration × dry mass; primary transport index (PTI) was calculated following Moral et al. (1994) as shootmetal concentrations/rootmetal concentrations.
After 20 days of normal culture, ecotypes Ph2-23 and Tor-1 were incubated with 10 μM CdCl2 for 30 min in a 4°C refrigerator or at room temperature (22°C), as reported previously (Luo et al., 2018). Their roots were then sampled and dried to constant weight. The Cd concentration was determined by ICP-MS after digestion with nitric acid as described above.
Extraction of Subcellular Components and Determination of Cd Concentrations
Twenty-day-old A. thaliana plants were treated with or without 10 μM CdCl2 for 4 days. The subcellular components were extracted by differential centrifugation (Weigel and Jäger, 1980). Briefly, fresh samples (0.5 g) were mixed with 8 mL extracting agent (250 mmol/L sucrose, 50 mmol/L Tris-HCl pH 7.5, 1 mmol/L dithiothreitol) and ground on ice to form homogenates. Each homogenate was centrifuged at 300 × g for 30 s to remove the residual CW fraction. Then, the samples were filtered and centrifuged at 20,000 × g for 45 min to precipitate the organelles. The extracted CWs and organelles were dried in an oven, digested in a 1:4 mixture (vol/vol) of HNO3 and HClO4, and diluted 100-fold with deionized water. The soluble fraction was again diluted 10-fold with deionized water, and Cd concentration was determined by ICP-MS.
Cell wall fractions were extracted as described by Brummell et al. (2004). The samples were quickly homogenized in 80% ethanol and incubated for 20 min at 90°C in a water bath. After cooling, the samples were centrifuged at 6000 × g for 10 min, and the precipitates were washed once with 1.5 mL 80% ethanol and 1.5 mL acetone (swirling for 2 min and centrifugation for 10 min at 6000 × g). Then, the supernatant was discarded, and the precipitate was soaked for 15 h in 1 mL dimethyl sulfoxide (for starch removal) and centrifuged for 10 min at 6000 × g. The supernatant was then discarded, and the CW precipitate dried for further use.
Determination of Cd in Pectin, Cellulose, and Hemicellulose
Pectin, cellulose, and hemicellulose were extracted according to Wu et al. (2017). The CW fraction was obtained from each sample and mixed with 5 mL 50 mM sodium acetate buffer containing 50 mM trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (CDTA) at pH 6.5. After shaking at 24°C for 12 h, the samples were centrifuged at 10,000 × g for 20 min. The residue was washed with 3 mL deionized water before centrifugation (5,000 × g, 10 min), and the supernatant comprised CDTA-soluble pectin. The residue was mixed with 5 mL 50 mM sodium carbonate solution, shaken for 12 h, and centrifuged at 10,000 × g for 20 min. The residue was washed with 3 mL water and centrifuged at 5,000 × g for 10 min. The supernatant comprised Na2CO3-soluble pectin. The residue was shaken with 5 mL 4 M KOH (containing 1% NaBH4) for 3 h and centrifuged at 5,000 × g for 20 min. The residue was washed with 3 mL water before centrifugation (5,000 × g, 10 min). The supernatant was the hemicellulose fraction, and the precipitate the cellulose fraction. After the cellulose precipitate was dried to a constant weight, 3 mL nitric acid was added for digestion. In parallel, 1 mL solution of CDTA-soluble pectin, Na2CO3-soluble pectin and hemicellulose, which was extracted previously, was digested with 3 mL nitric acid. The resulting solution was diluted 100-fold with deionized water. Metal concentrations were determined by ICP-MS with a NexIONTM 350X instrument (PerkinElmer).
Determination of Pectin Methylesterase (PME) Activity and Pectin, Cellulose, and Hemicellulose Concentrations
Pectin concentrations were detected using the Pectin Assay Kit (Shanghai Zcibio, Co., Ltd., China). Initially, each shoot sample (0.1 g) was homogenized by grinding with 5 mL distilled water, followed by low temperature centrifugation at 8000 × g for 10 min. Reagents I and II were incubated at 37°C for 10 min before beginning the assays. One hundred microliters of each sample was mixed with 100 μL reagent II. The control tubes contained 100 μL of each sample and 100 μL distilled water. The standard tube was prepared by mixing 100 μL reagent II with 100 μL reagent I, whereas the control tube was prepared by mixing 100 μL reagent II with 100 μL distilled water. The mixture of each tube was diluted to 800 μL with concentrated sulfuric acid, and the absorption at 530 nm was read after incubation at 95°C for 5 min in a water bath.
A Pectinesterase Assay Kit (Suzhou Comin Biotechnology, Co., Ltd., China) was used to detect PME activity as follows. Shoots (1 g/sample) were fully ground in 2 mL extraction reagent and centrifuged at 12,000 × g and 4°C for 15 min, after which each supernatant was transferred to a 15-mL centrifuge tube. Then, 50 μL reagent II and 8 mL reagent I were added, mixing after each addition, and then the pH was adjusted to 7.8 with reagent IV. Each centrifuge tube was placed in an oven at 37°C for 60 min, the pH was adjusted to 7.8 every 20 min, and the amount of reagent IV consumed was recorded. One unit of enzymatic activity (U) was defined as 1 μmol of NaOH consumed per minute by each tissue.
Cellulose concentrations were detected using a Cellulose Assay Kit (Beijing Solarbio Science & Technology Co., Ltd., China) and the extracted CW samples (approximately 5 mg each) as described by Brummell et al. (2004). The cellulose was homogenized with 0.5 mL distilled water, and the volume was adjusted to 0.5 mL. Then, each sample was placed in an ice-water bath, and 0.75 mL of concentrated sulfuric acid was added slowly, incubated for 30 min, and centrifuged at 8000 × g for 10 min. Then, 300 μL supernatant was mixed with 70 μL of a mixture of reagent I and reagent II, 630 μL concentrated sulfuric acid was added, and the resulting solution was incubated in a water bath at 95°C for 10 min, cooled to room temperature, and the absorbance measured at 620 nm.
Hemicellulose concentrations were detected using the Hemicellulose Assay Kit (Beijing Solarbio Science & Technology Co., Ltd.). Initially, 20-day-old A. thaliana plants were treated with or without 10 μM CdCl2 for 4 days. The shoots were dried to a constant weight, ground thoroughly, and passed through a 40-mesh sieve. After adding 1 mL 80% ethanol and vortexing, each sample was incubated in a water bath at 90°C for 10 min and centrifuged at 8,000 × g for 10 min. After washing each precipitate with distilled water, samples were dried to a constant weight. Then, 0.5 mL reagent I and 0.5 mL reagent II were added sequentially, and the samples were incubated in a water bath at 90°C for 1 h, mixed well, and centrifuged at 8,000 × g for 10 min. To each supernatant sample (125 μL) 125 μL reagent II and 750 μL distilled water were added, and the samples were incubated at 90°C for 5 min and cooled to room temperature before measuring their absorbance at 540 nm.
RNA Extraction and Determination of Gene Expression Levels
Cadmium-treated roots and shoot samples (0.2 g) were placed in liquid nitrogen, and total RNA was extracted with TRIzol (Ambion, Inc., Austin, United States). Complementary DNA (cDNA) templates were then synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China). Relative gene expression levels were determined by quantitative real-time PCR (qRT-PCR) using SYBR Premix Ex-TaqTM II (Takara Bio Inc., Kusatsu, Japan). The sequences of the primers used in the assays are shown in Supplementary Table S1, and the specific primers for the qRT-PCR analysis were designed using Primer Premier 6.0 software1.
Transcriptome Sample Preparation and Analysis
Shoots and roots of Ph2-23 and Tor-1 plants grown for 20 days and sampled after Cd treatment for 3 days were used for transcriptome analysis. We employed the mixed-sampling method (Kawahara et al., 2012), with each biological replicate included three samples of the same treatment. Biological replicates were quickly submerged in liquid nitrogen and sent to Gene Denovo Biological Sequencing Company (Guangzhou, China) for RNA-sequencing (RNA-Seq) analysis. We use R2 to carry out the Principal Component Analysis (PCA). Go function annotation of the identified proteins was carried out in GO database3. This process can be summarized as sequence alignment, mapping, calculation, hypergeometric tests, and annotation augmentation. The data are presented in a heat map, which was prepared using GraphPad Prism 84. The Venn diagram was generated using a publicly available website5.
Statistical Analyses
We performed minimum differential multiple-range comparisons to analyze our data, with each experiment being carried out with at least four biological replicates. P < 0.05 was considered to reflect a significant difference, and P < 0.01 was considered to reflect a highly significant difference. Analyses were conducted in SPSS software6. All bar charts were prepared with GraphPad Prism 8.
Results
Ecotype Tor-1 Coped Better With Cd Stress Than Ecotype Ph2-23
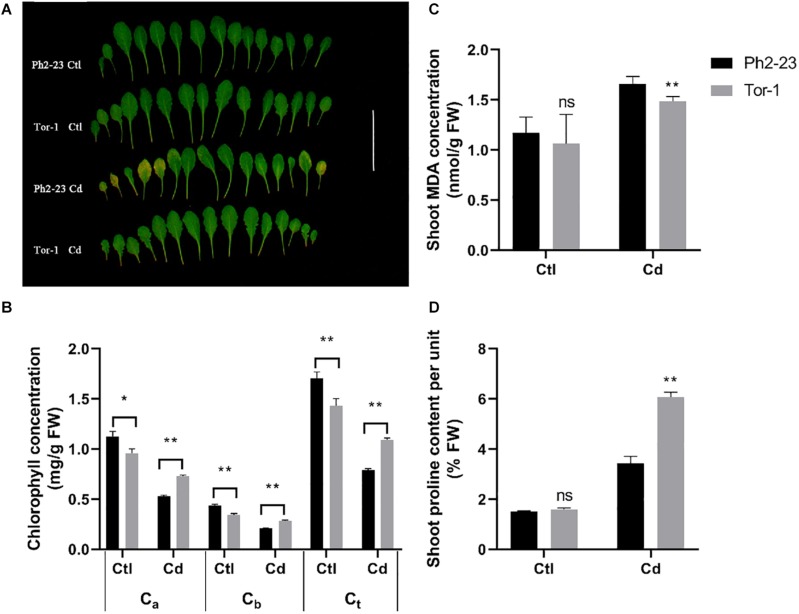
After 4 days of Cd treatment, the leaves of A. thaliana ecotype Ph2-23 became progressively more yellowish during Cd treatment, while no difference was found between the two ecotypes under control (without) Cd treatment (Figure 1A). Compared with those in Tor-1, the concentrations of chlorophyll a (Ca), chlorophyll b (Cb), and total chlorophyll (Ct) in Ph2-23 significantly decreased following exposure to Cd toxicity (Figure 1B). Under control conditions, the MDA concentration in the shoots of Ph2-23 and Tor-1 plants were similar, but after Cd treatment, the MDA concentration in Ph2-23 was significantly higher than that in Tor-1 (Figure 1C). After Cd treatment, the proline concentration in Tor-1 was significantly higher than that in Ph2-23, whereas no differences were observed between the two ecotypes under the control treatment (Figure 1D). Based on the observed phenotype and physiological indicators measurements, Tor-1 seems to cope better with the constraints of Cd exposure than Ph2-23.
FIGURE 1.
Differences in Cd tolerance of two Arabidopsis ecotypes treated with or without 10 μM CdCl2 for 4 days. (A) Photograph of leaves showing that Ph2-23 was more sensitive to Cd stress than Tor-1. (B) Chlorophyll loss under Cd2+ compared to control. Ca: Chlorophyll a; Cb: Chlorophyll b; Ct: total Chlorophyll. (C) Shoot malondialdehyde (MDA) concentration in the two ecotypes under control and Cd treatment. (D) Percentage of shoot proline content under control and Cd treatment. Ctl means control treatment, with normal culture conditions; Cd means treatment with CdCl2. Data are means of four measurements (n = 4), and the vertical bars indicate the SD. One (*) and two (**) asterisks indicate significant differences from the control at P < 0.05, and 0.01, respectively, ns, differences are not significant.
Ecotype Ph2-23 Translocated Less Cd to the Shoots Than Tor-1
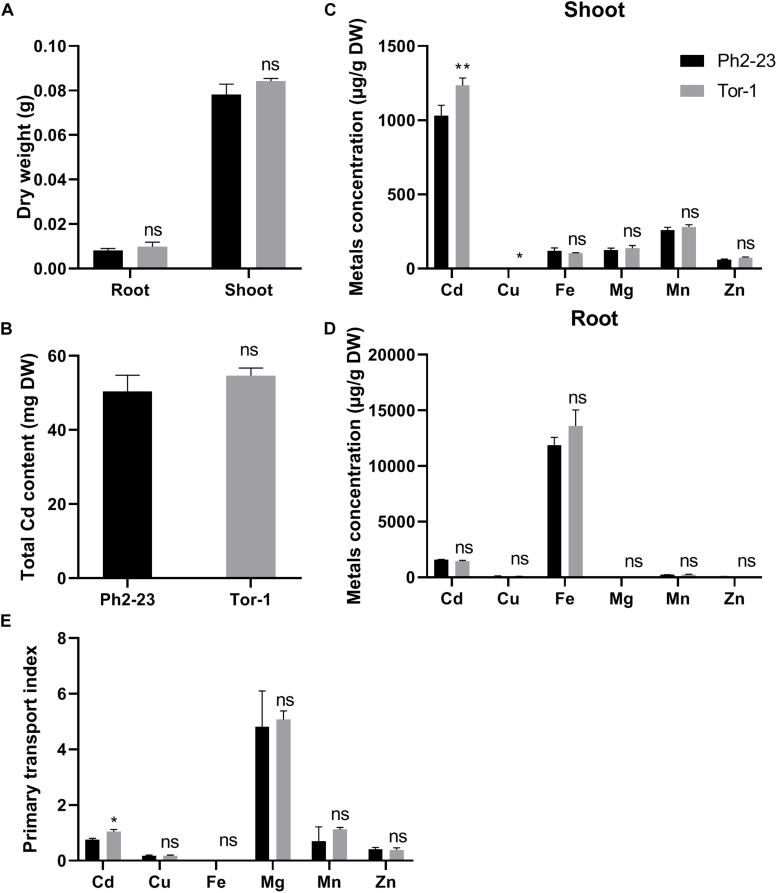
No significant difference was found in the dry weights of shoots and roots (Figure 2A) and total Cd concentration during Cd treatment between Ph2-23 and Tor-1 (Figure 2B). At both low (4°C) and normal (22°C) temperature treatments, the root absorption of Cd did not differ markedly between the two ecotypes after Cd treatment for 30 min (Supplementary Figure S1). The Cd concentration in the shoots of Tor-1 was significantly higher than that in the shoots of Ph2-23 (Figure 2C), but with no differences in roots (Figure 2D). In addition, no significant differences were observed in the concentrations of other metal elements (iron, magnesium, manganese, and zinc) between the shoots and roots of the two varieties, except for copper (Cu) concentration, which was lower in the shoots of Ph2-23 than that of Tor-1, while no differences were observed in root Cu concentrations. The primary transport index (PTI) of Cd in Tor-1 was higher than that in Ph2-23 but no differences were found between the two ecotypes in the PTI of other metals (Figure 2E). These data indicated that Tor-1 translocated more Cd to shoots, though was less affected by Cd stress.
FIGURE 2.
The uptake and distribution of Cd in two Arabidopsis ecotypes. (A) Effects on dry weight under 10 μM CdCl2 treatment. (B) Total Cd content in two ecotypes with or without 10 μM CdCl2 treatment for 4 days. (C) Shoot and (D) Root mentals distribution. (E) Primary transport index of metals (shootmetal concentrations/rootmetal concentrations). Data are means of four measurements (n = 4), and the vertical bars indicate the SD. One (*) and two (**) asterisks indicate significant differences from the control at P < 0.05, and 0.01, respectively, ns, differences are not significant.
Cd Distribution in Cellular Fractions of Ph2-23 and Tor-1
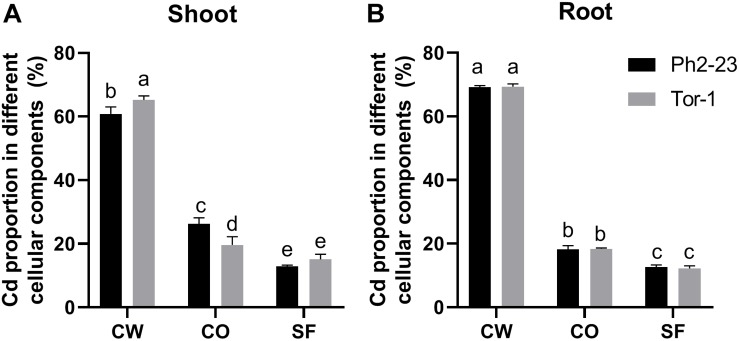
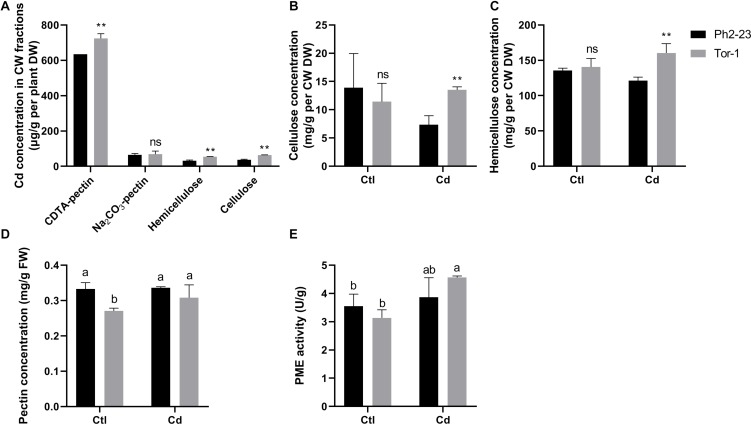
In both ecotypes, most Cd accumulated in the CW, followed by organelles, and then by the soluble fraction (Figures 3A,B). More Cd was detected in the CWs of Tor-1 shoots and less Cd was present in the organelles (Figure 3A). The Cd concentrations in the different components of root cells did not differ significantly between the two ecotypes (Figure 3B). This suggests that the relatively high concentration of Cd in the shoot CW may be responsible for the difference observed in Cd tolerance of these two ecotype. Shoot Cd concentrations in the CW CDTA-pectin, hemicellulose, and cellulose in Tor-1 were significantly higher than those in Ph2-23, while no difference in Na2CO3-pectin associated Cd concentration observed between the two ecotypes (Figure 4A). No obvious differences were observed for the cellulose content between the two ecotypes under control conditions. However, after Cd treatment, the cellulose concentration in Tor-1 became significantly higher than that of Ph2-23 (Figure 4B), and similar trend was observed for the hemicellulose concentration (Figure 4C). Pectin concentration in Tor-1 increased after Cd treatment, but not in Ph2-23 (Figure 4D). Similar results were also found for PME activity (Figure 4E).
FIGURE 3.
Distribution of Cd in the subcellular fractions of shoots (A) and roots (B) of the two Arabidopsis ecotypes grown under control or 10 μM CdCl2 treatment. CW, cell wall; CO, cellular organelles; SF, soluble fraction. Data are means of four measurements (n = 4), and different letters indicate means are significant differences (P < 0.05).
FIGURE 4.
Cd concentration and changes of cell wall components between Ph2-23 and Tor-1 with or without 10 μM CdCl2 treatment for 4 days. (A) Cd concentrations of CW components. (B–D) Cellulose, hemicellulose, pectin concentration in shoots. (E) PME activity in shoots. Ctl means control treatment, with normal culture conditions; Cd means treatment with CdCl2. Data presented are means of four measurements (n = 4), and different letters indicate means are significantly different (P < 0.05), two (**) asterisks indicate significant differences from the control at P < 0.01, ns, differences are not significant.
Differentially Expressed Genes (DEGs) Between Ph2-23 and Tor-1
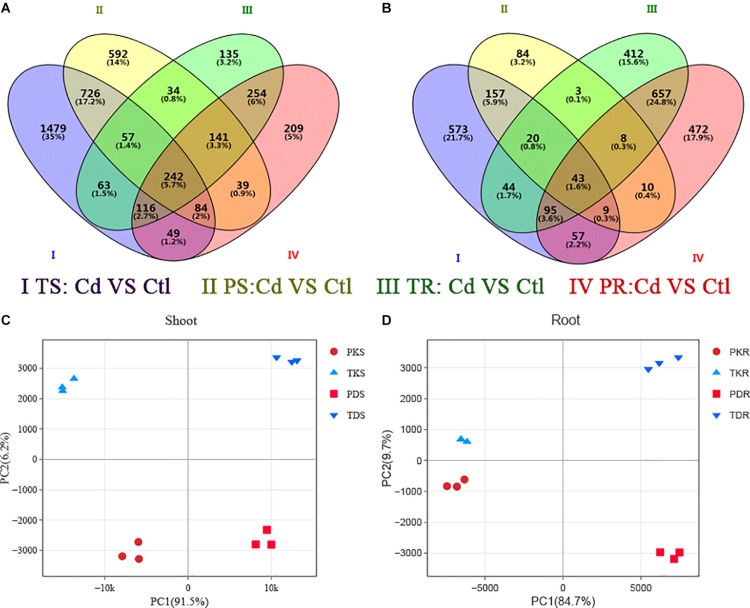
RNA-Seq analysis was performed to understand the molecular basis of the differential Cd sensitivity observed in the two Arabidopsis ecotypes. Compared to Ph2-23, Tor-1 showed more up-regulated genes (Figure 5A, Supplementary Figure S2, and Supplementary Table S2) and fewer down-regulated genes in shoots and roots following Cd exposure (Figure 5B, Supplementary Figure S2, and Supplementary Table S2). Both ecotypes shared 1109 up-regulated and 229 down-regulated Cd-responsive genes in the shoots (Figure 5A), and 753 up-regulated and 803 down-regulated Cd-responsive genes in the roots (Figure 5B). PCA showed that the first principal component (PC1) explained 95.1 and 84.7% while PC2 explained 6.2 and 9.7% of the variation in gene expression in the shoots (Figure 5C) and roots (Figure 5D) of the two cultivars, respectively.
FIGURE 5.
The transcriptome of differentially expressed genes (DEGs) in the two Arabidopsis ecotypes. Venn diagram analyses of (A) up-regulated and (B) down-regulated genes in two groups. P, Ph2-23; T, Tor-1; S, shoots; R, roots; K, control (Ctl); D, Cd. Data in each group are means of three replicates. Principal component analysis in shoot (C) and roots (D), Ph2-23 and Tor-1 sample distributions according to PC1 and PC2 are shown, with three biological replications, the thresholds for selecting DEGs were | log2 FC| > 1, FDR < 0.05.
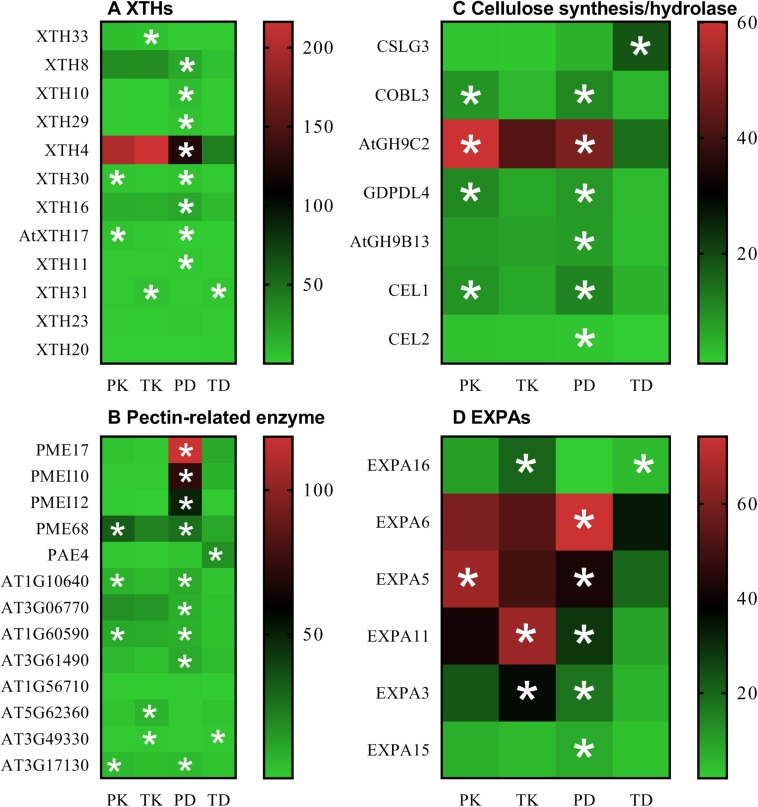
Gene Ontology (GO)-based enrichment analysis of the RNA-Seq data conducted on Tor-1 and Ph2-23 highlighted the DEGs representing enrichment of “CW” genes in the shoots (Supplementary Figure S3 and Supplementary Table S3). Taking into account the differences found in CW Cd accumulation and the GO enrichment analysis, we focused on the CW. We identified 60 DEGs (between Ph2-23 and Tor-1) involved in CW modification with a false-discovery rate (FDR) < 0.05. These DEGs encoded 31 proteins related to CW degradation, six proteins related to CW synthesis, and 22 other proteins involved in the CW (Supplementary Table S4). Genes related to CW degradation were more expressed in Ph2-23. In addition, Cd treatment induced 112 CW-related genes in Tor-1, compared with only 60 CW-related genes in Ph2-23 (Supplementary Table S4). Under Cd treatment, the expression of most XTHs, pectin lyases (AT1G10640, AT3G06770, AT1G60590, AT3G61490), PMEs (PME17, PME68), PMEIs (PME17, PMEI10, PMEI12), and one pectin methylesterase inhibitor superfamily protein (AT3G17310) were higher in Ph2-23 than in Tor-1 (Figures 6A,B and Supplementary Table S2); however, some pectin methylesterase inhibitor superfamily proteins (AT5G62360, AT3G49330) were significantly down-regulated after Cd treatment in Tor-1 (Figure 6B and Supplementary Table S2). The expression of CSLG3 (involved in cellulose synthesis) was up-regulated after Cd treatment in Tor-1 compared to Ph2-23 and the expression of COBL3 (also involved in cellulose synthesis) was higher in Ph2-23 with or without Cd treatment. Moreover, the expression levels of cellulose hydrolases (such as ATGH9C2, ATGH9B13, CEL1, and CEL2) were significantly lower in Tor-1 than in Ph2-23 (Figure 6C). We also found that the expression levels of EXPAs were higher in Ph2-23 than in Tor-1, except for EXPA16, which showed higher expression in Tor-1 under control treatment (Figure 6D).
FIGURE 6.
Transcriptome analysis of candidate DEGs under the control and Cd treatment in the two Arabidopsis ecotypes. (A) XTHs expression, (B) the gene expression of pectin-related enzyme, (C) the gene expression of cellulose synthesis/hydrolase, (D) EXPAs expression. PK and TK refer to Ph2-23 and Tor-1 under control treatment, respectively; PD and TD refer to Ph2-23 and Tor-1 following Cd treatment, respectively. Data presented means ± SE (n = 3). The thresholds for selecting DEGs were | log2 FC| > 1, FDR < 0.05, with one (*) asterisk indicates significant differences from the control at P < 0.05. Different colors indicate differential expressions, with red color indicating high expression, and green indicating low expression.
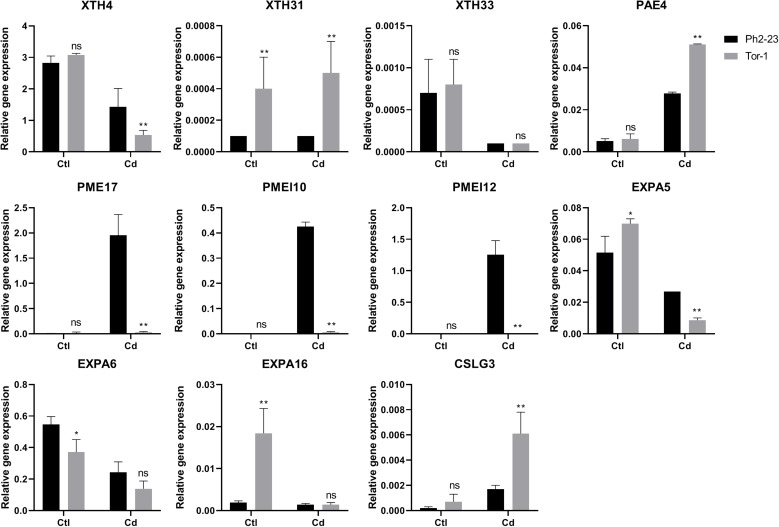
We verified the accuracy of the transcriptomics data through qRT-PCR, and the correlations between the RNA-Seq data and qRT-PCR data was 0.9517 (Figure 7 and Supplementary Figures S4A,B, S5). The qRT-PCR data also confirmed that XTH4, PAE4, PME17, PMEI12, PMEI10, EXPA5, EXPA6, and CALG3 were induced by Cd (Figure 7), consistent with our RNA-Seq data.
FIGURE 7.
qRT-PCR expression data of genes in shoots and roots between Ph2-23 and Tor-1. Ctl means control treatment, with normal culture conditions; Cd means treatment with CdCl2. Data are means of four measurements (n = 4), and the vertical bars indicate the SD. One (*) and two (**) asterisks indicate significant differences from the control at P < 0.05, and P < 0.01, respectively, ns, differences are not significant.
Discussion
In this study, we found that A. thaliana ecotype Tor-1 is more tolerant to Cd toxicity than ecotype Ph2-23, based on their phenotypes and the chlorophyll concentrations (Figures 1A,B). MDA is the main secondary product generated by the peroxidation of polyunsaturated fatty acids and it can reflect the degree of damage to cellular membranes (Del Rio et al., 2005). Furthermore, higher plants can accumulate proline in response to external stresses (e.g., heavy metals, drought, or high salt), which can be beneficial in mitigating these stresses (Fabro et al., 2004; Haudecoeur et al., 2009). Less cellular toxicity was observed in Tor-1 than in Ph2-23 (Figures 1C,D), which led us to further investigate which mechanism(s) are likely underlying the difference in tolerance observed between these ecotypes. There was no difference in dry weight between Tor-1 and Ph2-23 but the Cd concentration in the shoots of Tor-1 was significantly higher than that of shoots of Ph2-23, suggesting more Cd had accumulated in the shoots of Tor-1 than in those of Ph2-23 (Figures 2A,C, 4A). However, no significant difference was observed in the total Cd content between the two ecotypes (Figure 2B), suggesting that the difference in tolerance might be related to genes associated with Cd transport or cellular compartmentation.
Plant tolerance to Cd may be regulated by various genes responsible for Cd uptake and transport. ATP-binding cassette (ABC)-type transporters can transport more Cd–PC complexes to vacuoles in A. thaliana, which reduces the toxicity of Cd in the cytoplasm (Brunetti et al., 2015). In addition, Cd can be transported directly to vacuoles via calcium/hydrogen inhibitors (CAXs), metal-transporter proteins (MTPs), and heavy metal ATPase 3 (HMA3), which are tonoplast-localized transporters (Menguer et al., 2013; Pittman and Hirschi, 2016; Shafiq et al., 2019). In the present study, the expression of HMA2, responsible for Cd/zinc transport, was higher in Tor-1 than in Ph2-23, and CAX4 expression was higher in Tor-1 roots. Although more Cd might be transferred to vacuoles by CAX4 in Tor-1 roots than in Ph2-23 roots, Cd translocation from the roots to the shoots of Tor-1 was also higher (Figure 2E), and the expression levels of MTP1, HMA3, HMA4, ABCC1, and ABCC2 were not significantly different (Supplementary Figures S4A,B). These findings demonstrated that the differences in tolerance between Ph2-23 and Tor-1 were not primarily due to Cd-transport-related genes.
Previous reports showed that plants could chelate Cd through the CW during detoxification (Peng et al., 2017; Zhang et al., 2019). Cd can be fixed in CWs to reduce the toxicity of Cd to cellular organelles (Yu et al., 2019). Thus, we measured Cd concentrations in different cell fractions and found that the shoot CWs of Tor-1 contained more Cd, with less Cd allocated to organelles, and with no significant differences in roots (Figures 3A,B). Data from previous studies showed that exogenous auxin promoted A. thaliana hemicellulose synthesis, which reduces toxicity (Zhu et al., 2013b), and that the root CW polysaccharides of Sedum alfredii increased during Cd treatment, especially pectin and hemicellulose polysaccharides (Li et al., 2015). Further fractional extraction of the CW revealed that CDTA-pectin, cellulose, and hemicellulose fixed more Cd in Tor-1 (Figure 3C), suggesting that differences in their abundances in the CW may have caused the Cd concentration differences.
The degree of pectin acetylation can be regulated by PAEs, which may expose negatively charged groups, enabling them to bind Cd (Philippe et al., 2017). The demethyl esterification of A. thaliana pectin that can be modified by PMEs, PMEI was reported to inhibit PME activity (Röckel et al., 2008; Sénéchal et al., 2015), as was PME17 (Sénéchal et al., 2014). Pectin lyases are a group of enzymes that are thought to contribute to the degradation of pectin (Cao, 2012). Among the DEGs in the CW after Cd treatment, PAE4 expression was significantly higher in Tor-1; genes inhibiting PME activity were down-regulated in Tor-1, while genes inhibiting PME activity and pectin lyases were up-regulated in Ph2-23. The expressions of most PMEIs, as well as pectin lyases and PME inhibitor proteins, were significantly higher in Ph2-23 than in Tor-1. Furthermore, genes that inhibit PME activity were down-regulated in Tor-1 after Cd treatment, but not in Ph2-23 (Figure 6B). Increased pectin concentrations and PME activities were also found in Tor-1 compared with Ph2-23 (Figures 4D,E).
XTHs is known to be involved in hemicellulose synthesis in the primary CWs of higher plants and can play important roles in plant responses to metals such as aluminum and Cd (Zhu et al., 2013b; Han et al., 2014; Xuan et al., 2016). Our transcriptome analysis showed that XTH-expression levels were generally higher in Ph2-23 than in Tor-1 after Cd treatment, especially XTH4 (Figures 6A, 7). In addition, after Cd treatment, the hemicellulose content of Ph2-23 was significantly lower than that of Tor-1 (Figure 3D). Thus, Tor-1 may sequester Cd in the CW by increasing the hemicellulose content through XTHs under Cd treatment. Hemicellulose was widely viewed as a potential substrate for the production of liquid fuels and other value-added materials (Sun et al., 2012), meaning that using these XTH candidate genes to increase the hemicellulose content in plants with large biomass might not only help plants tolerate Cd toxicity but might also be used for increasing their production.
EXPAs may work in conjunction with cellulase and pectin lyase to break down CWs (Medina-Escobar et al., 1997; Llop-Tous et al., 1999; Harrison et al., 2001), although the exact mechanism(s) whereby EXPAs act on Cd is unclear. Previous findings showed that PtoEXPA12 over-expression (OX) increased Cd accumulation in tobacco plants (Zhang H. et al., 2018), although another study showed that TaEXPA2 OX is involved in tobacco resistance to Cd (Ren et al., 2018). After EXPA10 was knocked out, aluminum accumulation in the root hair region of rice plants decreased (Che et al., 2016). In the present study, EXPAs were significantly higher in Tor-1, except for EXPA16 (Figures 6D, 7); therefore, we hypothesized that EXPAs might be involved in Cd tolerance, although further testing is required to confirm this hypothesis.
Plant cellulose is produced by cellulose synthases, such as cellulose synthase-like (CSL) enzymes, members of the AtCesA cellulose synthase family, and members of the COBRA-LIKE (COBL) gene family (Manfield et al., 2004; Ben-Tov et al., 2015); cellulose hydrolysis in plants is regulated by endo-1,4-beta-glucanase (Levy et al., 2002). Less cellulose was detected in Ph2-23 after Cd treatment than in Tor-1 (Figure 3D). Although the expression of COBL2, which is involved in cellulose synthesis, was also higher in Ph2-23 than in Tor-1, expression of the cellulose synthase gene CSLG3 was much lower, and genes related to cellulose hydrolysis (such as AtGH9C2, AtGH9B13, and CEL1) were up-regulated (Figure 6C). These findings suggest that Tor-1 may regulate cellulose synthesis through these genes, leading to Cd binding. Cellulose is used in large quantities in fuel, and, like hemicellulose, whether these candidate genes can be transformed and/or overexpressed to enhance the synthesis of cellulose in plants with large biomass to achieve Cd tolerance and for cellulose production awaits further investigation.
Conclusion
Arabidopsis thaliana ecotype Tor-1 accumulated more Cd in the shoots and was more tolerant of Cd than A. thaliana ecotype Ph2-23. The difference in tolerance between these two ecotypes mainly resulted from variation in polysaccharide concentrations in the CW. The main anti-Cd toxicity mechanism of Tor-1 may involve gene regulation (e.g., XTH4, CSLG3, PME17, PAE4) to increase the cellulose and hemicellulose contents of the CW and the degree of pectin modification (to increase the CW retention of Cd), thereby reducing Cd toxicity in cellular organelles. Our study provides a strong evidence for the importance of shoot CW polysaccharides in Cd tolerance and identified candidate genes that could potentially be used to enhance Cd tolerance.
Data Availability Statement
The datasets generated for this study can be found in the NCBI using accession number PRJNA611457.
Author Contributions
YX and ZZ designed the experiments and analyzed the data. YX performed most of the experiments. YX, J-SL, AI, and ZZ wrote the manuscript. All authors contributed to the initial design of the project, read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was partly supported by the National Key R&D Program of China (2017YFD0200100 and 2017YFD0200103); National Natural Science Foundation of China (31800202); Province Key R&D Program of Hunan (2018NK1010); China Postdoctoral Science Foundation (2018M630900); Hunan Provincial Recruitment Program of Foreign Experts; the National Oilseed Rape Production Technology System of China; “2011 Plan” supported by The Chinese Ministry of Education; Double First-Class Construction Project of Hunan Agricultural University (kxk201801005); and the Innovative Research Groups of the Natural Science Foundation of Hunan Province (2019JJ10003).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00473/full#supplementary-material
References
- Åkesson A., Barregard L., Bergdahl I. A., Nordberg G. F., Nordberg M., Skerfving S. (2014). Non-renal effects and the risk assessment of environmental cadmium exposure. Environ. Health Perspect. 122 431–438. 10.1289/ehp.1307110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24:1. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39 205–207. 10.1016/j.dental.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M. A., Rager J. E., Smeester L., Fry R. C. (2011). Comparative genomic analyses identify common molecular pathways modulated upon exposure to low doses of arsenic and cadmium. BMC Genomics 12:173. 10.1186/1471-2164-12-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tov D., Abraham Y., Stav S., Thompson K., Loraine A., Elbaum R., et al. (2015). COBRA-LIKE 2, a member of the GPI-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiology 114:240671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell D. A., Dal Cin V., Crisosto C. H., Labavitch J. M. (2004). Cell wall metabolism during maturation, ripening and senescence of peach fruit. J. Exp. Bot. 55 2029–2039. 10.1093/jxb/erh227 [DOI] [PubMed] [Google Scholar]
- Brunetti P., Zanella L., De Paolis A., Di Litta D., Cecchetti V., Falasca G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66 3815–3829. 10.1093/jxb/erv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. (2012). The pectin lyases in Arabidopsis thaliana: evolution, selection and expression profiles. PLoS One 7:e46944. 10.1371/journal.pone.0046944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J., Yamaji N., Shen R. F., Ma J. F. (2016). An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 88 132–142. 10.1111/tpj.13237 [DOI] [PubMed] [Google Scholar]
- Chebli Y., Geitmann A. (2017). Cellular growth in plants requires regulation of cell wall biochemistry. Curr. Opin. Cell Biol. 44 28–35. 10.1016/j.ceb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Del Rio D., Stewart A. J., Pellegrini N. (2005). A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutrition 15 316–328. 10.1016/j.numecd.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Deng F., Yu M., Martinoia E., Song W. Y. (2019). Ideal cereals with lower arsenic and cadmium by accurately enhancing vacuolar sequestration capacity. Front. Genet. 10:322. 10.3389/fgene.2019.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H., Schaur R. J., Zollner H. (1991). Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11 81–128. 10.1016/0891-5849(91)90192-6 [DOI] [PubMed] [Google Scholar]
- Fabro G., Kovács I., Pavet V., Szabados L., Alvarez M. E. (2004). Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol. Plant Microbe Interact. 17 343–350. 10.1094/mpmi.2004.17.4.343 [DOI] [PubMed] [Google Scholar]
- Han Y., Sa G., Sun J., Shen Z., Zhao R., Ding M., et al. (2014). Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environ. Exp. Bot. 100 74–83. 10.1016/j.envexpbot.2013.12.021 [DOI] [Google Scholar]
- Harrison E. P., McQueen-Mason S. J., Manning K. (2001). Expression of six expansin genes in relation to extension activity in developing strawberry fruit. J. Exp. Bot. 52 1437–1446. 10.1093/jexbot/52.360.1437 [DOI] [PubMed] [Google Scholar]
- Haudecoeur E., Planamente S., Cirou A., Tannieres M., Shelp B. J., Morera S., et al. (2009). Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 106 14587–14592. 10.1073/pnas.0808005106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wu F., Wu S., Cao Z., Lin X., Wong M. H. (2013). Bioaccessibility, dietary exposure and human risk assessment of heavy metals from market vegetables in Hong Kong revealed with an in vitro gastrointestinal model. Chemosphere 91 455–461. 10.1016/j.chemosphere.2012.11.066 [DOI] [PubMed] [Google Scholar]
- Ismael M. A., Elyamine A. M., Moussa M. G., Cai M., Zhao X., Hu C. (2019). Cadmium in plants: uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 11 255–277. 10.1039/c8mt00247a [DOI] [PubMed] [Google Scholar]
- Jia H., Wang X., Dou Y., Liu D., Si W., Fang H., et al. (2016). Hydrogen sulfide-cysteine cycle system enhances cadmium tolerance through alleviating cadmium-induced oxidative stress and ion toxicity in Arabidopsis roots. Sci. Rep. 6:39702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian S. F., Luo J. S., Liao Q., Liu Q., Guan C. Y., Zhang Z. H. (2019). NRT1.1 regulates nitrate allocation and Cadmium tolerance in Arabidopsis. Front. Plant Sci. 10:384. 10.3389/fpls.2019.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., Oono Y., Kanamori H., Matsumoto T., Itoh T., Minami E. (2012). Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One 7:e49423. 10.1371/journal.pone.0049423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy I., Shani Z., Shoseyov O. (2002). Modification of polysaccharides and plant cell wall by endo-1, 4-β-glucanase and cellulose-binding domains. Biomol. Eng. 19 17–30. 10.1016/s1389-0344(02)00007-2 [DOI] [PubMed] [Google Scholar]
- Li T., Tao Q., Shohag M. J. I., Yang X., Sparks D. L., Liang Y. (2015). Root cell wall polysaccharides are involved in cadmium hyperaccumulation in Sedum alfredii. Plant Soil 389 387–399. 10.1007/s11104-014-2367-3 [DOI] [Google Scholar]
- Llop-Tous I., Domınguez-Puigjaner E., Palomer X., Vendrell M. (1999). Characterization of two divergent endo-β-1, 4-glucanase cDNA clones highly expressed in the nonclimacteric strawberry fruit. Plant Physiol. 119 1415–1422. 10.1104/pp.119.4.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. S., Huang J., Zeng D. L., Peng J. S., Zhang G. B., Ma H. L., et al. (2018). A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 9:645. 10.1038/s41467-018-03088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux A., Martinka M., Vaculík M., White P. J. (2013). Root responses to cadmium in the rhizosphere: a review. J. Exp. Bot. 62 21–37. 10.1093/jxb/erq281 [DOI] [PubMed] [Google Scholar]
- Maldonado-Mendoza I. E., Dewbre G. R., Blaylock L., Harrison M. J. (2005). Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene 345 191–197. 10.1016/j.gene.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Manfield I. W., Orfila C., McCartney L., Harholt J., Bernal A. J., Scheller H. V., et al. (2004). Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J. 40 260–275. 10.1111/j.1365-313x.2004.02208.x [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N., Cárdenas J., Moyano E., Caballero J. L., Muñoz-Blanco J. (1997). Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol. Biol. 34 867–877. [DOI] [PubMed] [Google Scholar]
- Memon A. R., Schröder P. (2009). Implications of metal accumulation mechanisms to phytoremediation. Environ. Sci. Pollut. Res. Int. 16 162–175. 10.1007/s11356-008-0079-z [DOI] [PubMed] [Google Scholar]
- Menguer P. K., Farthing E., Peaston K. A., Ricachenevsky F. K., Fett J. P., Williams L. E. (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J. Exp. Bot. 64 2871–2883. 10.1093/jxb/ert136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C. L., Juraniec M., Huguet S., Chaves-Rodriguez E., Salis P., Isaure M. P., et al. (2015). Intraspecific variability of cadmium tolerance and accumulation, and cadmium-induced cell wall modifications in the metal hyperaccumulator Arabidopsis halleri. J. Exp. Bot. 66 3215–3227. 10.1093/jxb/erv144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moral R., Gomez I., Pedreno J. N., Mataix J. (1994). Effects of cadmium on nutrient distribution, yield, and growth of tomato grown in soilless culture. J. Plant Nutr. 17 953–962. 10.1080/01904169409364780 [DOI] [Google Scholar]
- Nishizono H., Ichikawa H., Suziki S., Ishii F. (1987). The role of the root cell wall in the heavy metal tolerance of Athyrium yokoscense. Plant Soil 101 15–20. 10.1007/bf02371025 [DOI] [Google Scholar]
- Parrotta L., Guerriero G., Sergeant K., Cai G., Hausman J. F. (2015). Target or barrier? The cell wall of early-and later-diverging plants vs cadmium toxicity: differences in the response mechanisms. Front. Plant Sci. 6:133. 10.3389/fpls.2015.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. S., Wang Y. J., Ding G., Ma H. L., Zhang Y. J., Gong J. M. (2017). A pivotal role of cell wall in cadmium accumulation in the Crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant 10 771–774. 10.1016/j.molp.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Philippe F., Pelloux J., Rayon C. (2017). Plant pectin acetylesterase structure and function: new insights from bioinformatic analysis. BMC Genomics 18:456. 10.1186/s12864-017-3833-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman J. K., Hirschi K. D. (2016). CAX-ing a wide net: Cation/H+ transporters in metal remediation and abiotic stress signalling. Plant Biol. 18 741–749. 10.1111/plb.12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Álvarez R., Ortega-Villasante C., Álvarez-Fernández A., Del Campo F. F., Hernández L. E. (2006). Stress responses of Zea mays to cadmium and mercury. Plant Soil 279 41–50. 10.1007/s11104-005-3900-1 [DOI] [Google Scholar]
- Ren Y., Chen Y., An J., Zhao Z., Zhang G., Wang Y., et al. (2018). Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to Cd toxicity. Plant Sci. 270 245–256. 10.1016/j.plantsci.2018.02.022 [DOI] [PubMed] [Google Scholar]
- Richter J., Ploderer M., Mongelard G., Gutierrez L., Hauser M. T. (2017). Role of CrRLK1L Cell Wall sensors HERCULES1 and 2, THESEUS1, and FERONIA in growth adaptation triggered by heavy metals and trace elements. Front. Plant Sci. 8:1554. 10.3389/fpls.2017.01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röckel N., Wolf S., Kost B., Rausch T., Greiner S. (2008). Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 53 133–143. 10.1111/j.1365-313x.2007.03325.x [DOI] [PubMed] [Google Scholar]
- Sénéchal F., Graff L., Surcouf O., Marcelo P., Rayon C., Bouton S., et al. (2014). Arabidopsis PECTIN METHYLESTERASE17 is co-expressed with and processed by SBT3.5, a subtilisin-like serine protease. Ann. Bot. 114 1161–1175. 10.1093/aob/mcu035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénéchal F., Mareck A., Marcelo P., Lerouge P., Pelloux J. (2015). Arabidopsis PME17 activity can be controlled by Pectin Methylesterase Inhibitor 4. Plant Signal. Behav. 10:e983351. 10.4161/15592324.2014.983351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiq S., Zeb Q., Ali A., Sajjad Y., Nazir R., Widemann E., et al. (2019). Lead, Cadmium and Zinc phytotoxicity alter DNA methylation levels to confer heavy metal tolerance in Wheat. Int. J. Mol. Sci. 20:4676. 10.3390/ijms20194676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5:9. 10.1105/tpc.5.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Tian C., Diamond S., Glass N. L. (2012). Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in neurospora crassa. Eukaryot. Cell 11 482–493. 10.1128/EC.05327-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Zhang H. (2012). Cadmium toxicity: effects on cytoskeleton, vesicular trafficking and cell wall construction. Plant Signal. Behav. 7 345–348. 10.4161/psb.18992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Yang B., Wan H., Fang X., Yang C. (2018). The differences of cell wall in roots between two contrasting soybean cultivars exposed to cadmium at young seedlings. Environ. Sci. Pollut. Res. 25 29705–29714. 10.1007/s11356-018-2956-4 [DOI] [PubMed] [Google Scholar]
- Wang T., Hua Y., Chen M., Zhang J., Guan C., Zhang Z. (2018). Mechanism enhancing Arabidopsis resistance to cadmium: the role of NRT1.5 and proton pump. Front. Plant Sci. 9:1892. 10.3389/fpls.2018.01892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel H. J., Jäger H. J. (1980). Subcellular distribution and chemical form of cadmium in bean plants. Plant Physiol. 65 480–482. 10.1104/pp.65.3.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Riaz M., Yan L., Du C., Liu Y., Jiang C. (2017). Boron deficiency in trifoliate orange induces changes in pectin composition and architecture of components in root cell walls. Front. Plant Sci. 8:1882. 10.3389/fpls.2017.01882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., An L., Lu H., Zhu C. (2009). Exogenous nitric oxide enhances cadmium tolerance of rice by increasing pectin and hemicellulose contents in root cell wall. Planta 230 755–765. 10.1007/s00425-009-0984-5 [DOI] [PubMed] [Google Scholar]
- Xuan Y., Zhou Z. S., Li H. B., Yang Z. M. (2016). Identification of a group of XTHs genes responding to heavy metal mercury, salinity and drought stresses in Medicago truncatula. Ecotoxicol. Environ. Saf. 132 153–163. 10.1016/j.ecoenv.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Yu H., Guo J., Li Q., Zhang X., Huang H., Huang F., et al. (2019). Characteristics of cadmium immobilization in the cell wall of root in a cadmium-safe rice line (Oryza sativa L.). Chemosphere 241:125095. 10.1016/j.chemosphere.2019.125095 [DOI] [PubMed] [Google Scholar]
- Zhang J., Martinoia E., Lee Y. (2018). Vacuolar transporters for cadmium and arsenic in plants and their applications in phytoremediation and crop development. Plant Cell Physiol. 59 1317–1325. 10.1093/pcp/pcy006 [DOI] [PubMed] [Google Scholar]
- Zhang H., Ding Y., Zhi J., Li X., Liu H., Xu J. (2018). Over-expression of the poplar expansin gene PtoEXPA12 in tobacco plants enhanced cadmium accumulation. Int. J. Biol. Macromol. 116 676–682. 10.1016/j.ijbiomac.2018.05.053 [DOI] [PubMed] [Google Scholar]
- Zhang Z. H., Zhou T., Tang T. J., Song H. X., Guan C. Y., Huang J. Y., et al. (2019). Multiomics landscapes uncover the pivotal role of subcellular reallocation of cadmium in regulating rapeseed resistance to cadmium toxicity. J. Exp. Bot. 2019:erz295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. F., Lei G. J., Wang Z. W., Shi Y. Z., Braam J., Li G. X., et al. (2013a). Coordination between apoplastic and symplastic detoxification confers plant aluminum resistance. Plant Physiol. 162 1947–1955. 10.1104/pp.113.219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. F., Wang Z. W., Dong F., Lei G. J., Shi Y. Z., Li G. X., et al. (2013b). Exogenous auxin alleviates cadmium toxicity in Arabidopsis thaliana by stimulating synthesis of hemicellulose 1 and increasing the cadmium fixation capacity of root cell walls. J. Hazard. Mater. 263 398–403. 10.1016/j.jhazmat.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Zhu X. F., Shi Y. Z., Lei G. J., Fry S. C., Zhang B. C., Zhou Y. H. (2012). XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 24 4731–4747. 10.1105/tpc.112.106039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. F., Wan J. X., Sun Y., Shi Y. Z., Braam J., Li G. X., et al. (2014). XTH17 interacts with XTH31 to confer XET action and affect aluminum sensitivity in Arabidopsis. Plant Physiol. 114: 243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI using accession number PRJNA611457.