Fig. 1. Architectures of the substrate-bound LonENZ and substrate-free, LonOFF configurations.
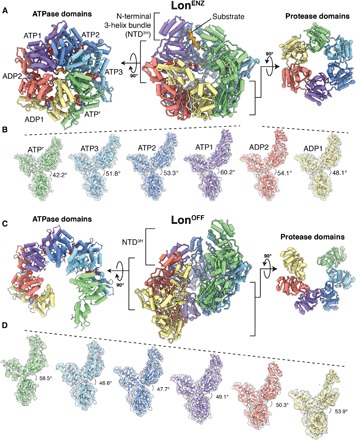
(A) Cutaway view of the substrate-bound Y. pestis LonENZ atomic model (center) flanked by orthogonal exterior views of the ATPase (left) and protease (right) domain rings. Each subunit of the homohexamer is assigned a distinct color depending on its position in the spiral staircase, and the cryo-EM density of the substrate is shown as a solid isosurface colored orange. Nucleotides are depicted using a sphere representation. (B) The different orientations of individual protomers relative to the protease domain, produced by orienting all the protease domains to a common view. Subunits are colored as in (A). The descending and ascending movements of the NTD3H and ATPase domains relative to their proteases are accentuated by dashed lines shown above the NTD3H. Dihedral angle measurements between ATPase and protease domains demonstrate a gradual expansion of descending subunits in the spiral staircase and compression in the two seam subunits. (C) Cutaway view of the substrate-free Y. pestis LonOFF atomic model (center) flanked by orthogonal exterior views of the ATPase (left) and protease (right) domain rings. Subunits are colored to correspond to their position in the LonENZ staircase architecture after transitioning. Nucleotides are depicted using a sphere representation. (D) The similar orientations of the individual protomers of the substrate-free structure are produced by orienting all the protease domains to a common view without changing axial position. The descending movement of the subunits is accentuated by dashed lines shown above the NTD3H. Dihedral angle measurements between ATPase and protease domains demonstrate subtle changes in the expansion and compression of the six descending subunits.
