Fig. 3. Lon proteolytic active site forms cleft for substrates in LonENZ and is autoinhibited in LonOFF.
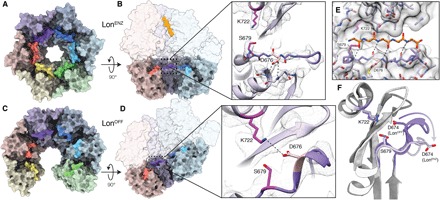
(A) An axial view of the chamber of the sixfold symmetric protease ring from the substrate-bound LonENZ structure is shown as a molecular surface with subunits are colored as in Fig. 1, with the six catalytic serine-containing loops (673 to 677) emphasized using a more saturated color. These loops organize into a ring-like assembly, generating a series of substrate-binding grooves. (B) A cutaway side view of the substrate-bound structure shown as a molecular surface. A close-up view of the cryo-EM density of the proteolytic active site is shown to the right as a gray mesh, with the atomic model showing the catalytic dyad (magenta) and serine-containing loop (purple stick representation). The serine-containing loop is likely stabilized in this extended conformation, in part, by hydrogen bonding between D676 (pink) and the peptide backbone of a nearby loop (light purple). (C) An axial view of the chamber of the protease ring from the LonOFF structure is shown and colored as in (A). The serine-containing loops are no longer interacting because of the separation of the protease domains in this conformation. (D) A cutaway side view of the substrate-free structure showing the location of the protease active sites in the open, exposed proteolytic chamber. A close-up view of the cryo-EM density of the proteolytic active site is shown as in (B), showing that in LonOFF, the catalytic serine-containing loop adopts a 310 helix that sterically occludes the proteolytic active site. D676 and K722 are within hydrogen-bonding distance, further limiting cleavage by the catalytic dyad. (E) A five–amino acid polyalanine peptide (orange) was modeled into the substrate-binding groove of LonENZ (represented using a transparent space-filling representation of the atomic model), based on the position of bortezomib covalently bound to S679 (PDB: 4YPM) (57). This demonstrates how an unfolded peptide substrate putatively docks into the active site for proteolytic cleavage by Lon protease. (F) The rearrangement of the serine-containing loop during the transition from LonOFF (dark gray/purple) to LonENZ (light gray/purple). This rearrangement is also shown in movie S2.
