Fig. 4. The PS1βH motif of Lon connects pore loop 1 to adjacent nucleotide-binding pockets.
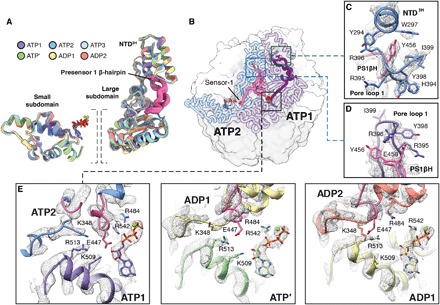
(A) Individual units of the AAA+ domains from the LonENZ structure are aligned and shown overlaid using a ribbon representation. The two units are (i) the small ATPase subdomains (left) and (ii) the NTD3H and large subdomain. Each subunit is colored according to the same color scheme assigned in Fig. 1. Superimposing these subdomains shows an average Cα RMSD of 0.81, indicating that these regions individually move as rigid units throughout the hydrolysis cycle. (B) ATP1 and ATP2 subunits are highlighted using a coil representation in the context of Lon structures (smoothed surface representation, ATPase domains are white, and protease domains are gray). The PS1βHs are colored purple and pink in ATP1 and ATP2, respectively. Sensor-1 at the C-terminal base of the PS1βH is denoted in ATP2. Close-ups of the regions enclosed by the boxes are shown in (C) and (D). (C) Y456, at turn of the PS1βH, stabilizes the NTD3H through proximal interactions with conserved aromatic residues Y294 and W297. (D) Pore loop 1 interactions with substrate are stabilized through interactions between E458 in the PS1βH and two tandem arginine residues in pore loop 1. (E) Remodeling cis and trans subunit interactions in the nucleotide-binding pocket drives sequential ATP hydrolysis cycle and stepwise substrate translocation. In the nucleotide-binding pocket of two adjacent ATP-bound subunits (e.g., ATP1 and ATP2; left), nucleotide is stabilized by interactions with the arginine finger (R484) in trans and sensor-2 (R542) in cis. Furthermore, a bridging glutamate residue (E447) at the N-terminal base of the PS1βH motif engages a cluster of basic residues within the nucleotide-binding pocket, stabilizing the intersubunit interface. In the ATP′ nucleotide-binding pocket (middle), this organization is disrupted upon E447 being retracted from the nucleotide-binding pocket upon nucleotide hydrolysis, causing the subunit to compress and bringing sensor-2 and other motifs involved in ATP hydrolysis (i.e., Walker A and Walker B motifs) in the proximity of the bound nucleotide. The ADP1 (right) and ADP2 (not shown) nucleotide-binding pockets reveal a similar organization to that observed in ATP′, indicating that nucleotide exchange is necessary to “reset” the hydrolysis cycle.
