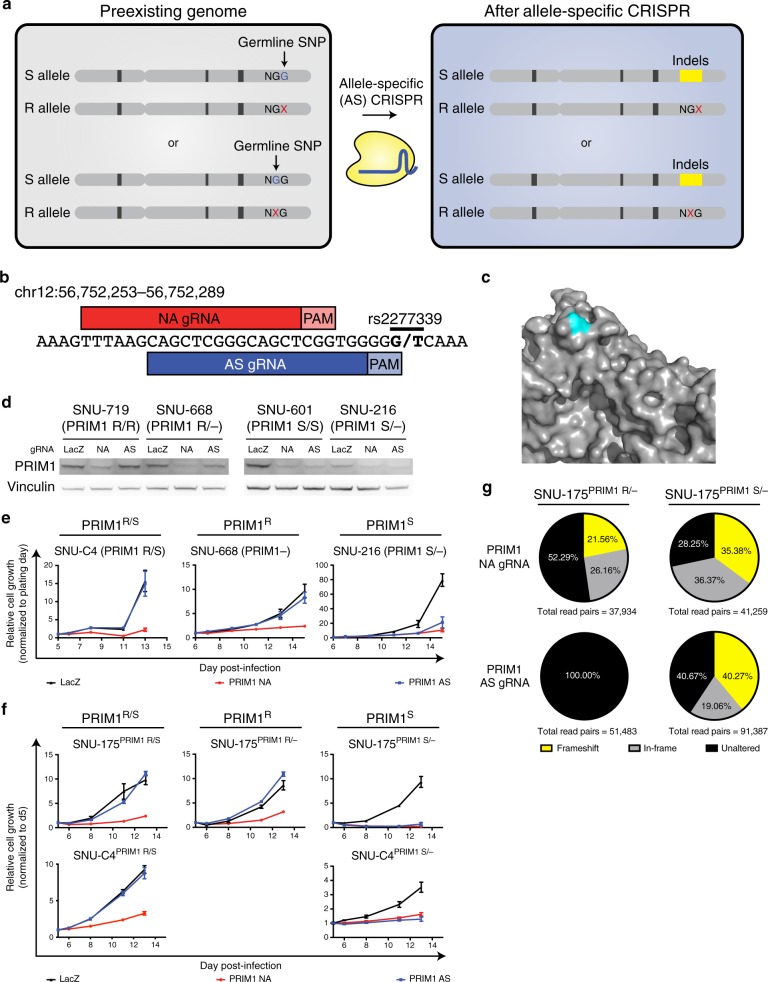
Fig. 2. Validation of PRIM1rs2277339 as a GEMINI vulnerability.
a Schematic indicating allele-specific CRISPR approach. “Preexisting genome” represents individuals heterozygous for a germline SNP in a S. pyogenes Cas9 protospacer adjacent motif (PAM) site. A “G” allele (blue) in the PAM retains Cas9 activity at the target site, making this allele CRISPR-sensitive (S). An allele other than “G,” represented by “X” (red) abrogates Cas9 activity at the target site, making this allele CRISPR-resistant (R). Expression of an allele-specific (AS) CRISPR sgRNA targeting the polymorphic PAM site leads to specific inactivation of the S allele. b Schematic of PRIM1 SNP rs2277339 locus showing target sites for positive control, non-allele specific (NA) sgRNA and experimental, allele-specific (AS) sgRNA. Alleles appear in bold. c Crystal structure of PRIM1 gene product88 shows the amino acid encoded by rs2277339 (teal) lies on the surface of the primase catalytic subunit (gray) near a potentially small-molecule accessible location. d Immunoblot of PRIM1 protein levels in indicated patient-derived cell lines expressing LacZ, PRIM1 NA, or PRIM1 AS sgRNA (n = 1 biological replicate). e Representative growth curves of indicated patient-derived cell lines expressing LacZ (black), PRIM1 NA (red), or PRIM1 AS (blue) sgRNA, as measured by CellTiter-Glo luminescence, relative to day of assay plating. n = 5 technical replicates. Data are presented as mean values ± s.d. See Supplementary Fig. 2 for additional biological replicates. f Representative growth curves of indicated isogenic cell lines expressing LacZ (black), PRIM1 NA (red), or PRIM1 AS (blue) sgRNA, as measured by CellTiter-Glo luminescence, relative to day of assay plating. n = 5 technical replicates. Data are presented as mean values ± s.d. See Supplementary Fig. 2 for additional biological replicates. g Disruption of PRIM1 in isogenic hemizygous PRIM1 resistant (PRIM1R) or PRIM1 sensitive (PRIM1S) cells expressing PRIM1 NA or AS sgRNA. Unaltered alleles (black), alleles with in-frame insertions or deletions (gray), and alleles with frameshift alterations (yellow) were assessed by deep sequencing of PRIM1 four days post-infection with sgRNA. Source data for Fig. 2d–g are provided as a Source Data file.