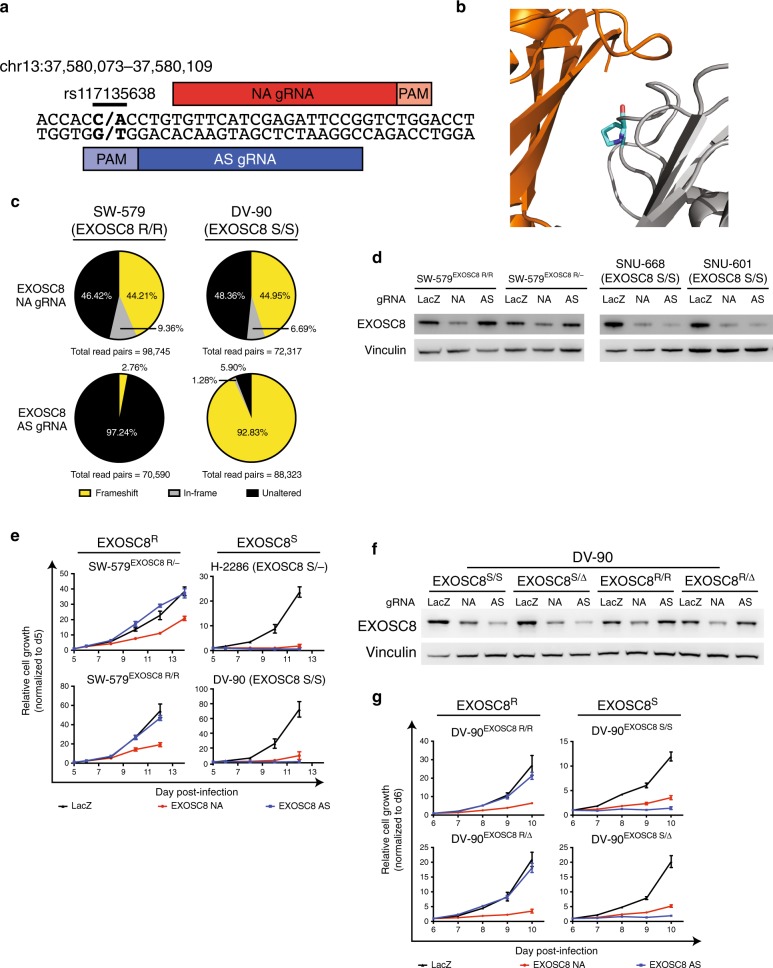
Fig. 3. Validation of EXOSC8rs117135638 as a GEMINI vulnerability.
a Schematic of EXOSC8 SNP rs117135638 locus showing target sites for positive control, non-allele specific (NA) sgRNA and experimental, allele-specific (AS) sgRNA. Alleles appear in bold. b Crystal structure of EXOSC8 gene product, Rrp4389 (gray) shows the amino acid encoded by rs117135638 (teal) lies on the surface of the Rrp43 protein near the interface with exosome complex subunit Mtr3 (orange). c Disruption of EXOSC8 in patient-derived EXOSC8 resistant (EXOSC8R) or EXOSC8 sensitive (EXOSC8S) cells expressing EXOSC8 non-allele specific (NA) positive control sgRNA or allele-specific (AS) experimental sgRNA. Unaltered alleles (black), alleles with in-frame insertions or deletions (gray), and alleles with frameshift alterations (yellow) were assessed by deep sequencing of EXOSC8 four days post-infection with sgRNA. d Immunoblot of EXOSC8 protein levels in indicated patient-derived and isogenic cell lines expressing LacZ, EXOSC8 NA, or EXOSC8 AS sgRNA (n = 2 technical replicates of 1 biological sample). e Representative growth curves of indicated patient-derived and isogenic cell lines expressing LacZ (black), EXOSC8 NA (red), or EXOSC8 AS (blue) sgRNA, as measured by CellTiter-Glo luminescence, relative to day of assay plating. n = 5 technical replicates. Data are presented as mean values ± s.d. See Supplementary Fig. 4 for additional biological replicates. f Immunoblot of EXOSC8 protein levels in indicated isogenic cell lines expressing LacZ, EXOSC8 NA, or EXOSC8 AS sgRNA (n = 2 technical replicates of 1 biological sample). g Representative growth curves of indicated isogenic cell lines expressing LacZ (black), EXOSC8 NA (red), or EXOSC8 AS (blue) sgRNA, as measured by CellTiter-Glo luminescence, relative to day of assay plating. n = 5 technical replicates. Data are presented as mean values ± s.d. See Supplementary Fig. 4 for additional biological replicates. Source data for Fig. 3c–g are provided as a Source Data file.