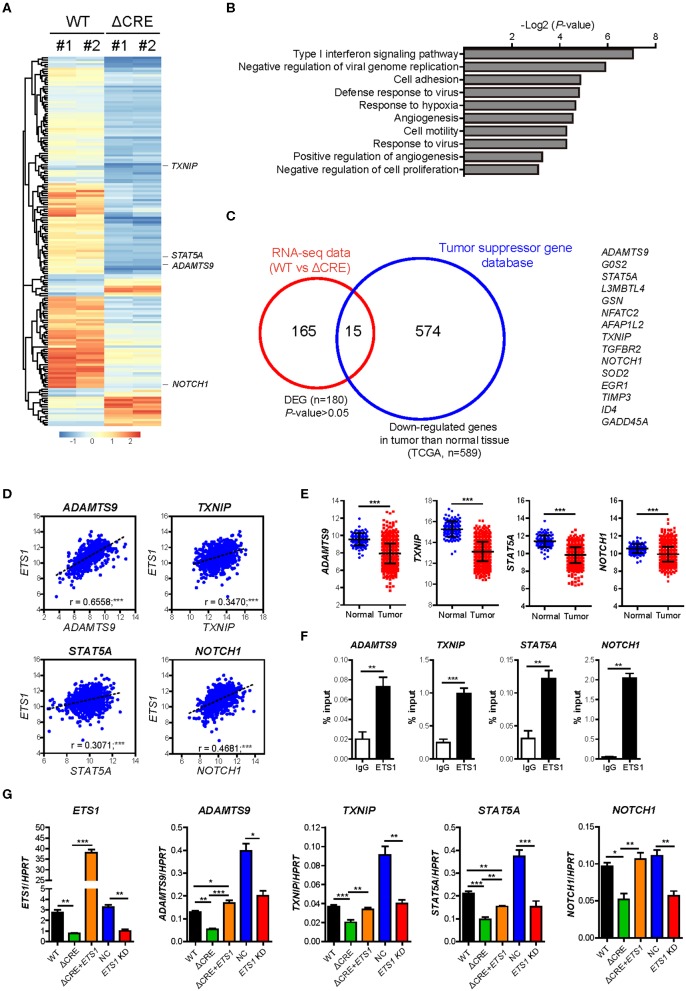
Figure 4.
ETS1 resultes muti-turmogeneic pathways in breast cancer cells. Transcriptome datasets from WT and ΔCRE cells were visualized by (A) heatmap analysis and further applied in (B) biological process of Gene Ontology (GO) enrichment analysis. (C) Venn diagram depicting the overlap of differentially expressed genes (DEGs) together with down-regulated genes in tumor compared to normal tissue, based on TSGene (33), a web resource for tumor suppressor genes. (D) Scatterplots depicting linear regression (black dot line) and Pearson correlation analysis with corresponding P-values. Correlation between ETS1 and target genes in BRCA. Each symbol represents an individual human specimen. (E) mRNA expression of ETS1 target genes in normal and tumor specimens. (F) ChIP assay was performed using anti-ETS1 antibody with MDA-MB-231 cells. Relative enrichments were determined by qRT-PCR with primers specific for ETS1 binding sites in the target gene locus. Representative data from three independent experiments. (G) Validation of representative target genes by qRT-PCR. Representative data from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (One-way ANOVA with Bonferroni test). *P < 0.05, **P < 0.01, ***P < 0.001 (Student t-test).