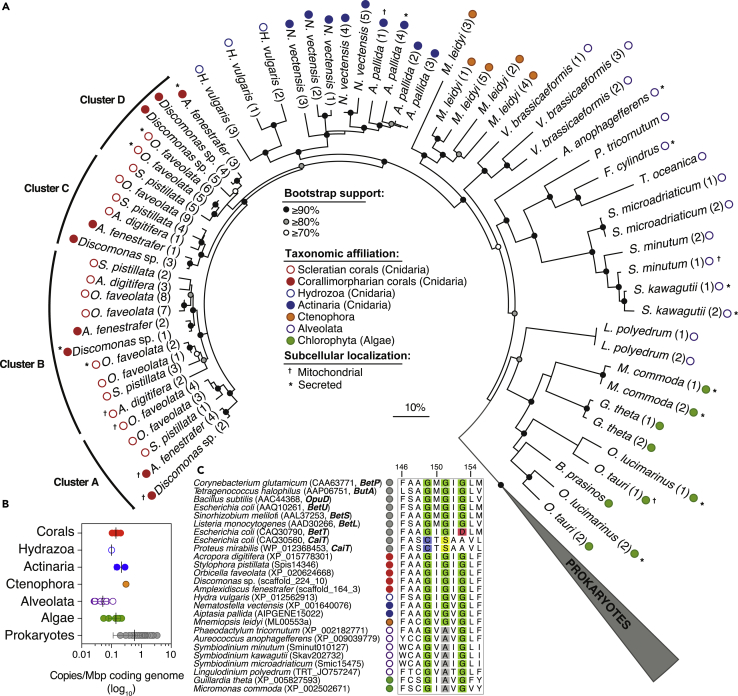
Figure 2.
Evolutionary History of Predicted Eukaryotic BCCT Carriers
(A) An unrooted maximum-likelihood phylogenetic tree showing the affiliation of predicted coral BCCT proteins (red circles) next to their closest cnidarian relatives (blue circles) and the basal prokaryotic BCCT carriers (wedged gray symbol). Colored circles highlight taxonomic affiliation of eukaryotic BCCT proteins. Note that the coral BCCTs are divided into four clusters (≥80% bootstrap support), with some putatively located in the mitochondria.
(B) The average density of predicted BCCT genes in marine invertebrates and marine prokaryotes with fully sequenced genomes (n = 125; Table S6); difference in copy number per mega-base pair (Mbp) of coding genome was only significant (one-way ANOVA, p = 0.0012) for Prokaryotes (gray circles) relative to Alveolata (green circles).
(C) Multiple aminoacid sequence alignment showing the G-x-G-x-G motif (highlighted in green) found in validated prokaryotic BCCT carriers specific for GB (BetP, ButA, OpuD, BetU, and BetL), choline (BetT), and carnitine (CaiT). Glycine residues lacking in choline and carnitine carriers are colored in red and blue (and yellow) respectively, whereas those missing in Symbiodiniaceae proteins are shown in gray. The amino acid positions reflect those in BetP from C. glutamicum. For brevity, only representative eukaryotic proteins are shown; Figure S6 provides the complete data.