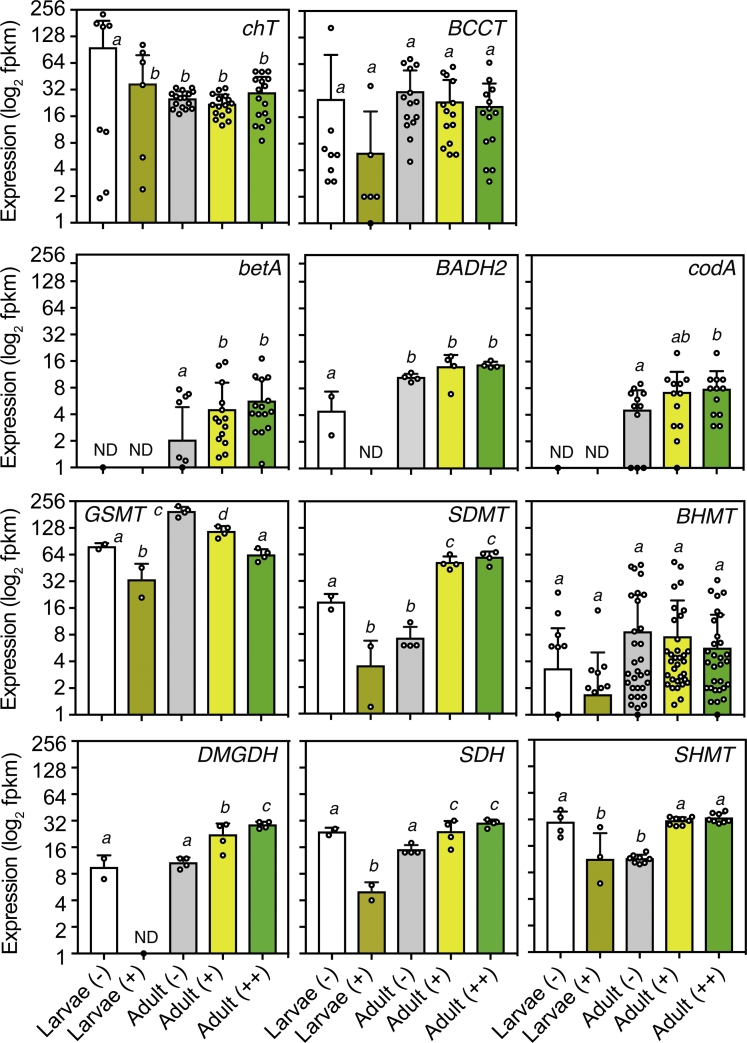
Figure 3.
Transcriptional Activity of GB Metabolism in the Coral Model Aiptasia
Gene expression was determined from published whole tissue transcriptomes (Baumgarten et al., 2015) of aquacultured Aiptasia larvae and adults fed regularly on brine shrimps (n = 2–4 replicates per experiment). Transcriptomes encompass various developmental and symbiotic states. Animals without dinoflagellate endosymbionts are indicated with “–”, whereas adults with intermediate or full endosymbiont levels are denoted with “+” and “++”, respectively. Barplots indicate average (± SD) expression in replicated samples and all predicted gene homologs (denoted by circular symbols). Statistically significantly different expression levels are denoted by different letters as deduced by one-way ANOVA tests (p > 0.05) with two-stage step-up Benjamin-Krieger-Yekutieli's multiple comparison test. ND, average expression was below ~1 FPKM.