Abstract
The traditional pipeline of hydrogel development includes individual one-by-one synthesis and characterization of hydrogels. This approach is associated with the disadvantages of low-throughput and high cost. As an alternative approach to classical one-by-one synthesis, high-throughput development of hydrogels is still tremendously under-represented in the field of responsive material development, despite the urgent requirement for such techniques. Here, we report a platform that combines highly miniaturized hydrogel synthesis with screening for responsive properties in a high-throughput manner. The platform comprises a standard glass slide patterned with 1 × 1 mm hydrophilic regions separated by superhydrophobic liquid-impermeable barriers, thus allowing deposition of various precursor solutions onto the hydrophilic spots without cross-contamination. The confinement of these solutions provided by the hydrophilic/superhydrophobic pattern allows encapsulation of cells within the hydrogel, and enables variation in hydrogel height and width. We have also proved the proper mixing of chemicals within the nanoliter-sized droplets. We have successfully implemented this platform for the synthesis of hydrogels, constructing 53 unique hydrogels, to demonstrate the versatility and utility of the platform. Photodegradation studies were performed on 20 hydrogels, revealing structure/function relationships between the hydrogel composition and photodegradability, and covering the range of degradability from non-degradable to rapidly degradable materials.
Keywords: High-throughput, Combinatorial libraries, Stimuli-responsive, Smart, Material, Microarrays
Graphical abstract
In order to discover and optimize stimuli-responsive materials fast, miniaturized high-throughput methodologies should be utilized. Herein, a droplet microarray platform for a nanoliter-scale high-throughput combinatorial hydrogel synthesis is demonstrated. The platform comprises hundreds of hydrophilic spots, serving as separate reaction microreservoirs, surrounded by superhydrophobic borders. Several libraries of photodegradable hydrogels are synthesized, covering the range of degradability from non-degradable to rapidly degradable materials.
1. Introduction
Hydrogels are widely applied in different biological and medical settings, including their use as matrices for three-dimensional (3D) cell culture, as drug delivery systems, and in regenerative medicine. Therefore, discovery of new hydrogel materials, as well as optimization of known materials to meet particular requirements, is of outstanding importance. Typically, hydrogels are synthesized and evaluated iteratively in a one-by-one manner. The drawback of this approach is the paucity of possible combinations that can be practically tested and the high cost in terms of time, consumables, and labor. Therefore, the process of discovery of novel hydrogels remains slow and the design principles, including structure-function relationships, are often not fully understood. Miniaturized high-throughput technologies, enabled by technologies such as droplet microfluidics [1] and polymer microarrays [2], are therefore important in facilitating and accelerating the discovery of novel hydrogels and screening of biological interactions of interest in parallel. Pioneered by Anderson et al. [3] and Tourniaire et al. [4] in the mid-2000s, polymer microarray technologies for biomaterial discovery have the advantage of an arrayed format. These techniques have been used successfully for the discovery of biomaterials for broad-ranging applications, ranging from materials for bone repair [5] and heart valve engineering [6], to materials for parasite removal [7] and bacteria-repellent medical devices [8]. High-throughput techniques have been widely used to study the effects of biomaterials on stem cells [[9], [10], [11]]. Biomaterial microarrays have been extensively reviewed [12]. Interestingly, reports of high-throughput synthesis of responsive materials are rare [13]. Responsive hydrogels have particular advantage of being synthesized in a high-throughput manner. Structure-function relationships between multiple responses cannot be easily forecast and thus, require experimental investigation in a process that does not lend itself to the classical iterative synthesis. The discovery of novel properties is facilitated by screening of a much larger range of chemical structures than can easily be achieved in conventional one-by-one synthesis. Therefore, high-throughput synthesis of responsive materials offers the potential to narrow the gap between the demand and supply of responsive materials.
Millions of cells are usually necessary to screen for cell-material interactions [3]. In the case of stem and primary cells, this can soon become expensive and therefore, restricts the number of possible combinations that can be tested. Solutions often have to be premixed before printing [3,14,15]. Furthermore, cell-material assays are usually performed without compartmentalization of the individual materials. Although understanding interactions of cells with biointerfaces is of utter importance [16,17], lack of compartmentalization can lead to cross-contamination issues in the case of cell-cell signaling studies, and is particularly important for the microenvironment of stem cells, thus demanding independent control of contact-mediated signaling. Introducing soluble factors after seeding cells can be conveniently achieved by compartmentalization. Compartmentalization also renders microarray add-ons [18] obsolete.
A platform using compartmentalized hydrophobic/hydrophilic patterns was developed by Le et al. [18], in which a pattern of varying wettability on gold-coated glass slides was used to study processes such as (human mesenchymal stem cells) hMSC encapsulation. However, the manual handling involved in this technique considerably limits the throughput. To the best of our knowledge, a platform that allows simple high-throughput fabrication of arrayed combinatorial library of multiple responsive hydrogels in separated nanoliter compartments has not yet been developed.
Recently, we established the droplet microarray (DMA) platform, a miniaturized platform based on arrays of nanoliter-sized droplets that can be used for high-throughput cell screening [19] and hydrogel microarray production [20]. The DMA platform comprises a standard microscopic glass slide photopatterned with hydrophilic regions/spots separated by superhydrophobic liquid-impermeable barriers. Following deposition of a unique prepolymerization mixture in each spot, these superhydrophobic barriers prevent cross-contamination, providing compartmentalization. Thus, the DMA platform can be used to perform multiple microbeads or cell encapsulation experiments in a parallel. Moreover, such assay miniaturization provides the ability to test a greater number of combinations of starting compounds, ensuring, if required, an almost spatially continuous gradient that can be used to study structure/function relationships. Specialized printing technique also allows precise deposition of compounds at the nanoliter level.
In this study, we demonstrated high-throughput combinatorial synthesis of cell-free hydrogels using DMA. A combinatorial library of smart hydrogels was synthesized on a DMA slide for direct investigation of the responsive properties of the hydrogels. Two combinatorial libraries of UV-responsive hydrogels (totaling 20 unique members á 16 replicates), all of which manifested a well-defined spatial gradient of components, were synthesized to highlight the convenience and future potential of DMA as a platform for responsive material synthesis.
2. Materials and methods
Poly(ethylene glycol)methyl ether methacrylate (PEGMA) (Mn approximately 500 g/mol), poly(ethylene glycol) dimethacrylate (PEGDMA) (Mn approximately 750 g/mol), 2-(N-3-sulfopropyl-N,N-dimethyl ammonium)ethyl methacrylate (SAMA), 2-(dimethylamino)ethyl methacrylate (DMAEMA), 2-butanone and D-(+)-glucose were purchased from Sigma–Aldrich (St. Louis, MI, USA). Ethyl (2,4,6-trimethylbenzoyl)phenylphosphinate was purchased from FluoroChem (Hadfield, Derbyshire, UK). Glucose oxidase (from Aspergillus niger) was purchased from VWR (Darmstadt, Germany). Sodium iodide was purchased from Fluka (Munich, Germany). DMA glass slides were purchased from Aquarray GmbH (Eggenstein-Leopoldshafen, Germany) and used without further changes.
2.1. Synthesis of photoinitiator sodium phenyl-2,4,6-trimethylbenzoylphosphinate
Ethyl (2,4,6-trimethylbenzoyl)phenylphosphinate (1 g, 0.003 mol) was dissolved in 10 ml 2-butanone. After the addition of sodium iodide (0.6 g, 0.004 mol), the reaction mixture was stirred overnight at 60°C. The precipitate was collected by filtration, washed with 2-butanone and dried. Sodium phenyl-2,4,6-trimethylbenzoylphosphinate (yield: 0.7 g, 79%) was yielded as a white solid. The identity of the product was confirmed by NMR:
1H-NMR (400 MHz, D2O): δ/ppm = 7.78–7.48 (Haromatic, phenyl), 6.92 (s, Haromatic, benzoyl), 2.27 (s, para-CH3), 2.06 (s, ortho-CH3) (Fig. S1).
2.2. Printing procedures
Monomers were deinhibited before use by passing over a short column of basic aluminum oxide (Alfa Aesar, Ward Hill, MA, USA). All printing experiments were performed using non-contact liquid dispensing technology (immediate drop on-demand technology (I-DOT) liquid dispenser; Dispendix, Stuttgart, Germany). The viscosity of aqueous monomer stock solutions was adjusted for reproducible printing by non-contact liquid dispensing technology. Printing of solutions was calibrated, allowing precise dosage of solutions in the nanoliter range. The humidity was maintained at 70% during printing.
2.3. Confirmation of adequate mixing of components in droplets
Even distribution of two monomers within droplet after printing was confirmed by fluorescence, Raman spectroscopy measurements and multiple particle tracking (MPT).
2.3.1. Fluorescence measurements
PEGMA solution was supplemented with rhodamine B and the solution of DMAEMA as an example monomer, and the second monomer was supplemented with fluorescein. The solutions of PEGMA, DMAEMA, sodium phenyl-2,4,6-trimethylbenzoylphosphinate (SAP), and PEGDMA (5 mol%) were then printed sequentially in quadruplicate. PEGMA was printed in descending volumes (60–10 nL; 12 dilutions), and DMAEMA was printed in ascending volumes (10–60 nL; 12 dilutions), and SAP and PEGDMA were printed in constant volumes (30 nL). The homogeneity of fluorescein and rhodamine distribution was assessed qualitatively by fluorescence microscopy (Keyence), and the mean intensity was determined via ImageJ (100 × 100-pixel square sections in the middle of each droplet).
2.3.2. Raman spectroscopy measurements
Raman spectroscopy measurements were obtained from a composition comprising 30 nL SAP, 60 nL PEGMA, 10 nL DMAEMA, and 30 nLl PEGDMA (5 mol%) with a Bruker Senterra spectrometer (Bruker Optics, Germany). For focusing of the excitation laser and collimation of backscattered light an Olympus MPLAN 20 × objective (NA 0.4) was used, resulting in a spot diameter of 5 μm on the sample surface. The excitation laser (λ = 532 nm) was operated at 10 mW output power. Each spectrum was integrated over 60 s with three coadditions (3 × 20 s). The total number of measurement spots per hydrogel micropad was 144, arranged in a 12 × 12 matrix (Fig. S4).
2.3.3. Multiple particle tracking
To determine the microrheological properties of the hydrogel, an exemplary composition comprising 30 nL SAP, 60 nL PEGMA, 10 nL DMAEMA, and 30 nL PEGDMA (5 mol%) was polymerized. SAP solution contained glucose oxidase and glucose and was further supplemented with green fluorescent polystyrene microspheres of diameter 0.5 μm (Bangs Laboratories, USA) as tracers, from which the Brownian motion was tracked. Then the resulting particle trajectories were transformed into mean square displacement (MSD) traces and quantitative information about the rheological properties were obtained based on a relation between the MSD as a function of lag time τ and the macroscopic complex shear modulus G∗(ω) as a function of the frequency ω [21]. The Laplace transform of the particle MSD is related to the complex modulus G∗ of the sample via a generalized Stokes-Einstein equation [22].
where a is the radius of the embedded beads, kB is the Boltzmann constant and T is the temperature. In addition, to perform the statistical analysis and characterize the microstructure heterogeneity, we examined the distribution of displacements (Van Hove correlation function) and calculated [23] the non-Gaussian parameter α:
This parameter is zero for a Gaussian distribution, expected for a homogeneous, uniform sample, while deviations from this distribution result in large α values, reflecting the presence of heterogeneities.
The experimental set-up was based on an inverted fluorescence microscope (Axio Observer D1, Carl Zeiss, Germany) equipped with a Fluar 100 × objective (numerical aperture 1.3 ×, 100 × magnification, oil immersion lens). Two-dimensional (2D) images (field of view 127 × 127 μm, frame rate 50 f/s) of these fluorescent beads were recorded using a sCMOS camera Zyla X (Andor Technology). The videos of the fluctuating microspheres were analyzed using the software Image Processing System (iPS, Visiometrics, Germany) and a self-written Matlab code [24], based on the widely used Crocker and Grier tracking algorithm [25].
2.4. Confinement studies
Owing to compartmentalization, different volumes of precursors can be deposited onto hydrophilic pattern of the DMA. To highlight this fact, 240 resp. 120 nl of prepolymerization mixture were printed onto the 1 × 1 mm patterned DMA, subsequently polymerized and incubatied in rhodamine B solution for better visualization. In addition, 40 nL of prepolymerization mixture was deposited onto the 500 × 500 μm patterned DMA, subsequently polymerized and incubatied in rhodamine B solution for better visualization.
To show the possibility of cell encapsulation within the hydrogels, a suspension of HeLa cells (10 nL, 5∙106 cells/ml) was printed directly after the hydrogel precursors. After the polymerization (360 nm, 6 mW/cm2, 10 min) cells were stained with 1 μg ml−1 Hoechst 33342 by immersing the DMA slide in the Hoechst stain solution for 20 min.
2.5. Construction of libraries
For optimization of the degassing procedure, a 36-member DMAEMA/PEGMA library (ratios varying from 6:1 to 1:6) with 2.5, 5.0, and 10.0 mol% PEGDMA was synthesized in quadruplicates. We varied the GOx concentration from 1 μM to 40 μM. After the polymerization and incubation in rhodamine B solution, the hydrogel widths were measured manually. After optimization, a typical printing procedure was performed using SAP solutions supplemented with glucose oxidase (40 μM) and glucose (0.1 M) and glucose oxidase solution being freshly prepared for every experiment, SAP/glucose oxidase/glucose solution being freshly prepared immediately before printing. All solutions were prepared with PBS and reactions were typically performed at room temperature.
In a typical printing procedure, hydrogel libraries were printed as follows: printing of 30 nL SAP (2.6 mg/ml in PBS) supplemented with glucose oxidase and glucose; overprinting with PEGMA (40 wt% in PBS) with the volume descending from 60 to 10 nL per spot; overprinting with 30 nL PEGDMA (e.g. 77,3 mg/ml in PBS for 5 mol% w.r.t. monomers), overprinting with SAMA (230 mg/ml in PBS) with the volume ascending from 10 to 60 nL per spot. To prevent evaporation, humidifying rows consisting of only 130 nL PBS per spot were printed so that each spot containing prepolymerization mixture was surrounded either by another prepolymerization mixture spot or by PBS spot. The DMA produced in this way was irradiated for 10 min at 6 mW/cm2 in a closed petri-dish containing cellulose pads soaked in water to provide a humid environment and washed with PBS. Some starting material combinations could not be polymerized due to both low cross-linker and PEGMA concentration and were not considered further. All degradation studies were carried out using 270 nm UV light source (UVACUBE 2000, Dr. Hönle AG, Gräfelfing, Germany) with irradiance of 22 mW/cm2.
3. Results and discussion
3.1. General array production
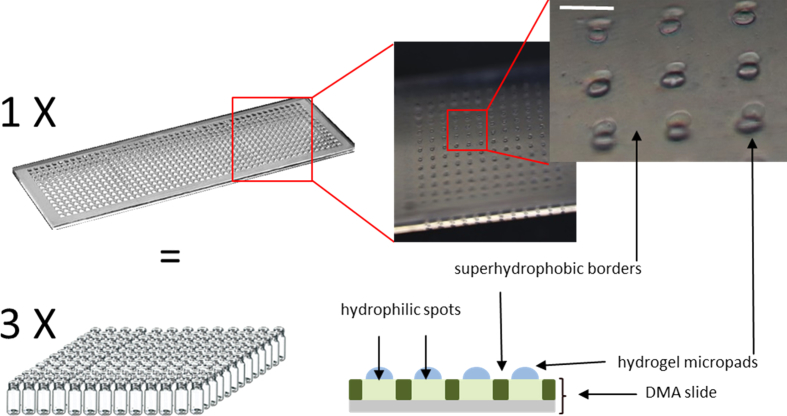
In this study, we aimed to develop a platform in which hundreds of highly adjustable combinations of polymerization starting materials and additives can be simultaneously synthesized and assayed. To implement the combinatorial approach and support a high-throughput aspect, we used a liquid-dispensing device based on non-contact liquid-dispensing technology to dispense droplets containing components of a hydrogel library [26]. The I-DOT printer is a non-contact printer that uses compressed air pulses to dispense precise dosage of solutions in the nanoliter range. The non-contact liquid-dispensing technology printer was equipped with a humidifier, thus allowing printing of aqueous solutions without adding humectants or oil films. Droplets can be dispensed directly onto the hydrophilic spots of the DMA without cross-contamination because superhydrophobic borders provide confined surroundings to hydrophilic regions. These wall-less microreactors allow synthesis of a unique material in each spot. The non-contact liquid-dispensing technology printer facilitates the creation of a typical library of 144 unique prepolymerization mixtures within several minutes. This process requires less than 20 μL of reagent solution in total and is executed in more than 800 pipetting steps. To construct a similar library in a conventional one-by-one manner on 1 ml scale would be much more time-consuming and require approximately 7000 times more solution (144 ml). An example of the material microarray first printed as a prepolymerization mixture and then subsequently photopolymerized and dried is shown in Fig. 1. All probed hydrogel micropads stayed firmly attached to the DMA surface after being immersed into aqueous media for at least one week.
Fig. 1.
Schematic comparison of high-throughput screening of 588 hydrogels in vials and on DMA. Confined hydrophilic regions on a DMA slide act as wall-less reaction microvessels, enabling the single DMA slide to substitute 588 vials. Scale bar 1 mm. DMA, droplet microarray.
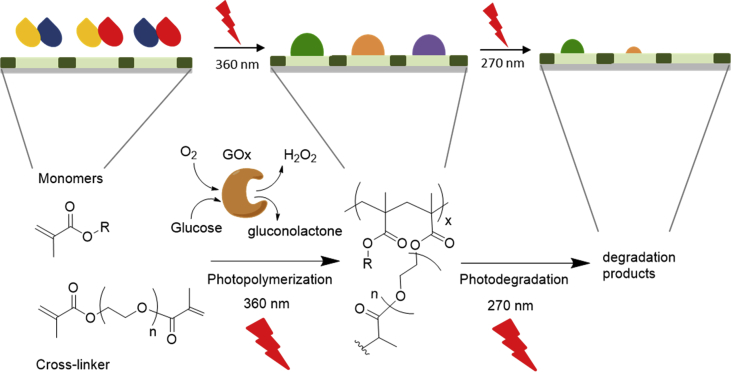
Recently, we have shown that commonly used methacrylate-based hydrogels are inherently degradable by 270 nm UV light [27]. Most of the high-throughput smart material libraries reported to date are based on temperature and water responses [13]; therefore, we aimed to complement these studies with a study of methacrylate-based photoresponsive hydrogels (Fig. 2). We selected photopolymerization because it is a fast, non-contact, controllable (in terms of wavelength, exposure time, and intensity), proceeds under physiological conditions and under high spatiotemporal control [28]. We used SAP as a photoinitiator; therefore, photopolymerization occurred at 360 nm and photodegradation occurred at 270 nm.
Fig. 2.
Schematic description and reaction scheme of high-throughput on-chip photoinduced synthesis of combinatorial library of photodegradable hydrogels and assessment of degradation properties. Based on this reaction, we have synthesized three libraries: 3-membered PEGMA/PEGDMA library, 33-membered PEGMA/DMAEMA/PEGDMA library, and 17-membered PEGMA/SAMA/PEGDMA library; PEGMA/DMAEMA/PEGDMA library was used to optimize the photopolymerization protocol; PEGMA/PEGDMA and PEGMA/SAMA/PEGDMA libraries were assessed with regard to their photodegradation properties. PEGMA, poly(ethylene glycol)methyl ether methacrylate; PEGDMA, poly(ethylene glycol) dimethacrylate; SAMA, 2-(N-3-sulfopropyl-N,N-dimethyl ammonium)ethyl methacrylate; DMAEMA, 2-(dimethylamino)ethyl methacrylate.
3.2. Polymerization on a nanoliter scale
Despite many benefits, the open infrastructure of DMA involves the contact of droplets containing prepolymerization mixture with oxygen, which can dramatically decrease the efficiency of radical polymerization. Conventional methods of oxygen removal, such as degassing via nitrogen purging, working under an inert atmosphere or freeze-pump-thaw cycles, require additional infrastructure or are not readily applicable to reactions carried out in submicroliter volumes [29]. Therefore, to perform miniaturized HT radical polymerization under ambient conditions and overcome its inherent intolerance to oxygen, we supplemented the initiator solution with glucose oxidase (GOx) and glucose (Glu), which function in combination as an oxygen scavenger system [[29], [30], [31]]. We used a PEGMA/DMAEMA library as a sample library for optimization of the degassing procedure. Without the GOx/Glu degassing system, the average hydrogel micropad size decreased as the concentration of DMAEMA increases (Fig. S2). For these compositions, the conversion was low due to presence of oxygen, resulting in insufficient (partial) cross-linking of polymers, which were consequently unable to adhere to the underlying substrate and were removed during washing step. For the GOx/Glu degassed polymerization, 40 μM GOx was sufficient to reproducibly synthesize hydrogel microarrays with an average hydrogel micropad size of 123 ± 14 μm in all prepolymerization mixtures tested. The high surface-to-volume ratio of submicroliter droplets combined with the shorter pathways for oxygen diffusion, as well as the suboptimal reaction temperature for GOx justifies the increased (200 × ) GOx concentration required for complete conversion of monomers within the hundred nanoliter-sized droplets compared to reactions conducted in the submilliliter range [32].
3.3. Confinement
For inkjet printing on flat surfaces with homogeneous wettability, the size of hydrogels is controlled by changing the number of droplets printed per spot. In contrast, the size and shape of the hydrogel features printed on DMA can be controlled by changing the freely selectable pattern of the photomask used for production of slides (Fig. 3 A). Prepolymerization mixtures deposited onto different patterns of choice can be therefore polymerized into arbitrary shapes and with freely selectable density of the hydrogel micropad owing to precise setting of the pitch between spots. Superhydrophobic borders prevent liquid-spreading and therefore, allow deposition of volumes up to 1000 nL in a 1 × 1 mm spot. Deposition of different volumes provides control over the hydrogel height and width, which would not be possible on a non-patterned surface (Fig. 3 A).
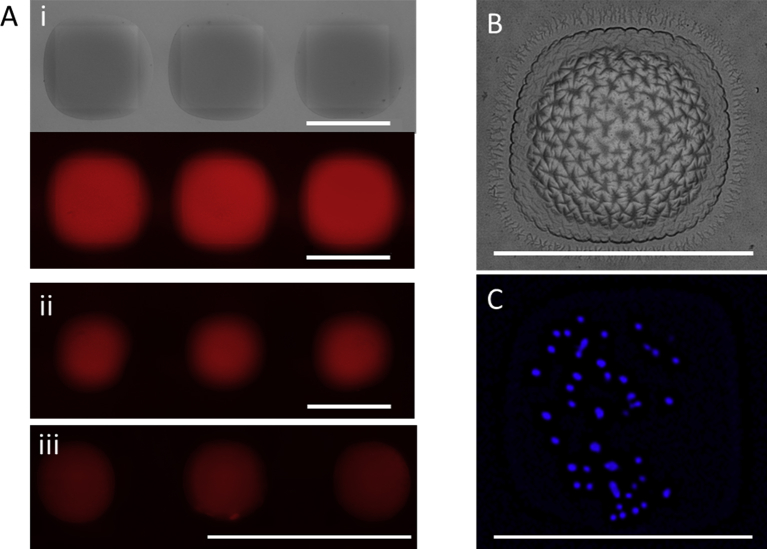
Fig. 3.
Confinement on DMA. (A) Brightfield and fluorescent images of hydrogels, which were polymerized in nanoliter-sized compartments using (i) and (ii) 1000 μm squared pattern, 240 nl, respectively. 120 nl prepolymerization mixture (iii) 500 μm squared pattern, 40 nl prepolymerization mixture. (B) Exemplary hydrogel showing surface instabilities upon extensive swelling. (C) HeLa-cells encapsulated within the hydrogel pad and stained with Höchst. Scale bars are 1 mm. DMA, droplet microarray.
Furthermore, in the absence of an oxygen scavenger system during the polymerization, we observed surface instabilities because of the oxygen gradient and hence, different degrees of crosslinking from the surface of the hydrogel to its core (Fig. 3 B). Because the hydrogel micropads are laterally confined on rigid hydrophilic spots, hydrogels may become unstable upon swelling in water and develop various surface patterns. Therefore, combinatorial synthesis of hydrogels using DMA can be used for high-throughput assessment of thin film buckling properties depending on the starting composition.
Owing to the compartmentalization, various objects can be encapsulated within the hydrogels, for example, magnetic beads, tracer particles (e.g. for microrheological measurements), and cells. We therefore demonstrated the possibility of encapsulation of cells within hydrogels (Fig. 3C) by overprinting the prepolymerization solution with a cell suspension. Confinement shows the possibility to study cell encapsulation parameters (rapid optimization of hydrogel composition to promote a certain biological outcome), to develop 3D cell culture matrices, as well as to study cell-material interactions without cross-talk between cells encapsulated in neighboring hydrogels prevented by superhydrophobic borders.
3.4. Mixing within droplets
During the printing process, the droplet of one solution impacts on the droplet of another solution to induce a vortex, which enables fluid mixing. To ensure the reaction occurs throughout the whole reaction volume, the mixing behavior of the reagents in consecutively printed droplets was evaluated. The printed solutions were colored one-by-one with fluorescein/rhodamine B and printed on top of each other in different proportions, one colored solution at a time, as described in the literature [33]. The overprinting of colorless droplet with a colored one (and vice versa) induced significant turbulence in the spots, resulting in even distribution of the fluorescent dye (Fig. S3) in the typical range of droplet volumes (60 nL–10 nL). The mixing time was estimated to be several seconds.
We also performed Raman spectroscopy measurements on one selected hydrogel composition to confirm the homogeneous distribution of chemicals within droplets. As shown in Fig. 4 B and S4, the distribution of C-H (3015–2790 cm−1), CH3 and CH2 (1500–1400 cm− 1), as well as carbonyl vibrations (1730–1700 cm−1) was radially even. The higher intensity of vibrations toward the top of the hydrogel micropad correlated with its height and the overall bulged form, and thus with the abundance of corresponding functional groups. The consistency of bulge formation was confirmed by white light scattering measurements (Fig. S4).
Fig. 4.
Proving proper mixing within the droplet on DMA. (A) Raman spectrum of one point of the hydrogel reveals the carbonyl and C–-H vibrations. (B) Raman mapping of C-H vibration on a representative hydrogel; (C) Mean square displacement values of tracer particles, and elastic and viscous moduli for three regions of the hydrogel (i) central region, 55 tracer particles; (ii) left region, 43 tracer particles, and (iii) bottom region, 62 tracer particles. DMA, droplet microarray.
Finally, we characterized rheological and heterogeneity properties of the hydrogel micropad performing MPT microrheology. To incorporate polystyrene tracer particles into the hydrogel before polymerization, we printed the SAP/glucose oxidase/glucose solution containing particles instead of SAP solution. After polymerization, the MSD of the tracer was measured. Fig. 4C shows the variation of MSDs as a function of lag time (τ) for polystyrene tracer particles dispersed in hydrogel in three different regions (i: central, ii: left, iii: bottom). In all cases, all MSDs exhibited almost no time dependence with slope close to zero and an average MSD (red curve) value approximately 2.10−4 μm2. This result indicates that particles are highly constrained by the surrounding fluid, which is consistent with elastic trapping of particles in a gel-like network. In addition, statistical analyses of the MSD distribution clearly reveals a homogeneous structure on the micrometer length scale with a small value of the non-Gaussian parameter α = 0.55, 0.56, and 0.41 determined at 0.1 s for central, left, and bottom region of the hydrogel micropad, respectively [23]. These results are also reflected in the variation of elastic and viscous moduli, G′ and G'´, respectively, calculated from the average MSD. As shown in Fig. 4C, independent of the region, a significant degree of elasticity was observed with G′(ω) > G′′(ω) and constant elastic modulus G′ = 14 ± 4 Pa in a broad frequency range. From this value, we can directly determine the mesh size of the network in accordance with the classical theory of rubber elasticity with and we found ξ = 67 ± 6 nm. Overall, fluorescence, Raman and MPT microrheological analyses confirmed even distribution of printed solutions in nanoliter-sized droplets analogous to the synthesis on a macroscale.
3.5. Combinatorial screening of hydrogels
To confirm the applicability of the DMA platform for synthesis and assessment of the properties of responsive hydrogels, we synthesized a proof-of-concept library with one photodegradable (methacrylate-based) and one non-degradable (acrylate-based) hydrogel. Both hydrogels were printed on the DMA in a checkerboard pattern, which could not be recognized before irradiation; all hydrogels had identical appearance (Fig. 5 Ai). Given an inherent photodegradability of polymethacrylates and the UV transparency of water, the remarkable minute-scale degradability of polymethacrylate-based hydrogels originates from the high swellability of the hydrogel network. Therefore, during UV irradiation, macroradicals are separated in space, and recross-linking is suppressed. In the case of polyPEG-acrylates, recross-linking reactions occur much faster than that of polyPEG-methacrylates, rendering acrylate-based hydrogels non-degradable under UV light, and allowing visualization of the hidden checkerboard pattern under UV irradiation (Fig. 5 Aii). This proof-of-concept library, comprising one degradable and one non-degradable material, arranged in a preprogrammed pattern, represents a special case of temporal and spatial control over the degradation. Furthermore, high-throughput combinatorial synthesis can be used to rapidly customize the hydrogel degradation properties for each specific application.
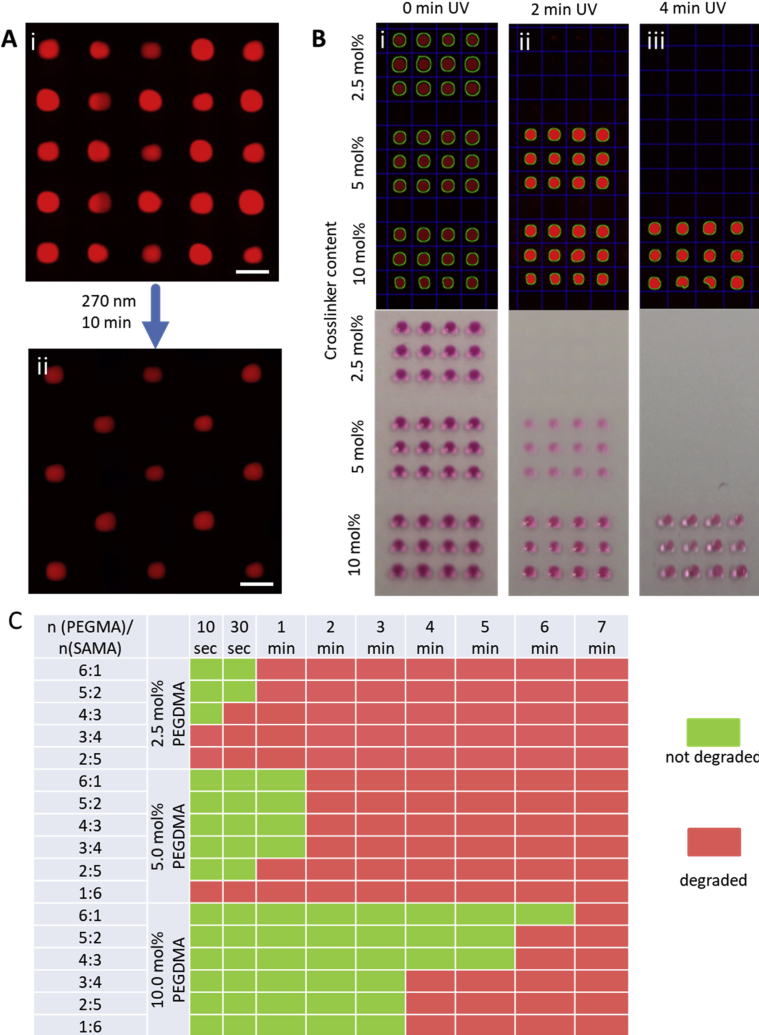
Fig. 5.
A) (i) Exemplary library of polyPEG acrylate and polyPEG methacrylate hydrogels arranged in a checkerboard-pattern on a microarray, colored with Rhodamine 6G; (ii) after illumination with UV light (270 nm, 22 mW/cm2, 10 min) only non-degradable polyacrylates are left. Scale bars are 1 mm. (B) Different degradation ability of PEGMA-based hydrogels with varying amount of cross-linker. Degradation can be followed either by software (top) or by eye (bottom); (i) before UV; (ii) 2 min UV (270 nm, 22 mW/cm2); (iii) 4 min UV (270 nm, 22 mW/cm2). (C) Photodegradation screening results of a two-dimensional combinatorial library, constructed by varying the amount of cross-linker PEGDMA and PEGMA/SAMA molar ratio. The degradation ability is inversely proportional to the amount of PEGDMA and PEGMA. PEGMA, poly(ethylene glycol)methyl ether methacrylate; PEGDMA, poly(ethylene glycol) dimethacrylate; SAMA, 2-(N-3-sulfopropyl-N,N-dimethyl ammonium)ethyl methacrylate.
Because the proof-of-concept library showed the possibility to distinguish between degradable and non-degradable hydrogels (yes/no), we next assessed hydrogels with different degradation times (therefore different degradability) on the DMA slide. As already mentioned, the swellability of the hydrogel network is the prerequisite for hydrogel photodegradation. Swellability correlates negatively with the amount of cross-linker, rendering hydrogels with higher cross-linker content (with respect to the monomer) less swellable, and therefore less degradable, compared to a hydrogel with a lower cross-linker content. To determine whether this behavior could be reproduced on the DMA, we synthesized a library of three PEGMA-based hydrogels with different cross-linker concentrations (2.5, 5.0, and 10.0 mol% w.r.t. PEGMA) (Fig. 5 B). To verify the reproducibility of the degradation, we synthesized each composition in 16 replicates, resulting in 48 nL-sized hydrogels that required only 6 μL of solution in total. After the synthesis, the hydrogel microarray was immersed in rhodamine 6G solution to color the hydrogels and was subsequently subjected to UV irradiation (270 nm, 1–3 min). The course of the degradation (i.e. the presence or disappearance of hydrogel micropads) was followed either by eye (pink rhodamine) or under a microscope in the fluorescent channel. If detection was made via fluorescence, hydrogel circles were detected automatically using specialized software. As expected and shown in Fig. 5 B, hydrogels with 2.5 mol% cross-linker were completely degraded within 2 min of UV irradiation, whereas doubling the cross-linker content slowed degradation of the hydrogels to 4 min. Hydrogels with the highest cross-linker content did not degrade even after 10 min UV irradiation. It is noteworthy that all 16 replicates of one composition degraded simultaneously, emphasizing the reproducibility of the synthesis.
Finally, we examined the effect of supplementation of the prepolymerization mixtures from the previous library with different amounts of zwitterionic SAMA on degradation of the resulting hydrogels. To do so, we varied the volume ratios of equimolar monomer solutions PEGMA:SAMA from 6:1 to 1:6 with PEGDMA concentrations 2.5, 5.0, and 10.0 mol% w.r.t. the combined amount of monomers, maintaining a constant ratio Σn(monomers)/n(cross-linker) for all compositions. Therefore, we extended the combinatorial library to another dimension, yielding 17 unique combinations with a total of 16 replicates each, resulting in 272 nL-sized hydrogels that required only 34 μL of solution in total. SAMA is a charged monomer that is known to increase the swelling ratio of hydrogels; therefore, we investigated the ability of increased SAMA:PEGMA ratios to increase the rapidity of hydrogel degradation on the DMA [22]. Two trends were observed following exposure of the DMA slide with SAMA library to UV light. Generally, as we have already shown, the rate of hydrogel degradation correlates negatively with the degree of cross-linking. In addition, clear trends were observed within the group of hydrogels with the same cross-linker content. As expected, progressively substituting PEGMA with SAMA rendered the hydrogels more degradable (Fig. 5C). Four compositions (PEGMA:SAMA = 6:1 and 4:3 with cross-linker concentrations 2.5 and 10.0 mol% each) were upscaled from 130 nL to 180 μL to verify the reproducibility of the synthesis. It is noteworthy that the synthesis of four hydrogel compositions in triplicate at a submilliliter scale required 2.16 ml prepolymerization solutions, which is sufficient to synthesize more than 17,000 hydrogels on a DMA. After illumination of upscaled hydrogels with UV for 1.5 min, the mass loss was measured. For 2.5 mol% cross-linker, the rate of (PEGMA:SAMA 6:1)-hydrogel degradation was slower than that of the (PEGMA:SAMA 4:3)-hydrogel (58% vs. 88%). Both upscaled hydrogels with 10.0 mol% cross-linker showed almost no degradation after 1.5 min UV (1% vs. 6% for 6:1 and 4:3 PEGMA:SAMA proportions, respectively). This also reflects the hydrogel behavior on the DMA, with hydrogels with 10.0 mol% cross-linker still visible after 1.5 min UV exposure. Therefore, high SAMA content and low cross-linker content were shown to correlate with high degradability on both the microscales and nanoscales.
4. Conclusions
The DMA platform meets the requirements for a cell-compatible polymer microarray [3] and can be used as a benchtop-ready platform for high-throughput combinatorial hydrogel synthesis and, if desired, subsequent cell encapsulation by providing multiple compartmentalized sites in hydrophilic spots in a hydrophilic-superhydrophobic pattern. To prove the general suitability of the platform to synthesize and evaluate hydrogels, we focused on studying the structure-function relationships of cell-free photoresponsive hydrogels. An array of confined nanoliter-sized droplets containing prepolymerization mixture was created within minutes by non-contact liquid dispensing and was subsequently photopolymerized. Using this approach, 20 unique hydrogel micropads á 16 replicates were produced. A spatial gradient of both PEGMA and SAMA monomers as well as cross-linker PEGDMA was produced by dispensing different volumes of these compounds along the x and y axes of an array. The photodegradation properties of the underlying materials were examined in a high-throughput manner, embracing the spectrum from non-degradable to rapidly degradable hydrogels and emphasizing the suitability of DMA to quickly assess structure/function relationships. These relationships allow for synthesis of hydrogels with defined degradation behavior, which can be used as, for example, dynamic sealants, and for controlled inanimate cargo release [27]. To use photodegradable materials for cell release, a cytocompatible strategy, possibly with less energetic wavelengths, should be established.
Spatially resolved libraries have an advantage of simple decoding and allow synthesis of numerous different polymers without the need of their separation [34]. High-throughput synthesis and screening can give quick insights into the structure/function relationships, enabling a customized variation of degradation kinetics by the choice of particular hydrogel precursors. Importantly, the temporal and spatial control over the degradation allows facile one-step on-demand formation of arbitrary hydrogel patterns by UV irradiation.
Because the number of printable additives is immense, one can cover a large combinatorial space through combinatorial high-throughput screening over a multitude of hydrogel prepolymerization mixtures (including, for example, peptides that promote cell adhesion, fibronectin, laminin, hyaluronic acid, dextran, chitosan to identify biomaterials). This approach will facilitate fine-tuning of the biochemical and/or biophysical properties of hydrogels and therefore, identification of optimal candidates for a certain application such as recapitulation of the complexity of extracellular matrix to optimize materials for cell encapsulation. We also advocate the use of high-throughput experimentation in the field of responsive hydrogels to generate new and more sophisticated responsive materials with novel properties, which may remain undiscovered using the one-by-one synthesis approach.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
CRediT authorship contribution statement
Alisa Rosenfeld: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Project administration, Validation, Writing - original draft. Claude Oelschlaeger: Investigation, Data curation, Formal analysis, Writing - original draft. Richard Thelen: Investigation, Data curation, Formal analysis, Writing - review & editing. Stefan Heissler: Investigation, Data curation, Formal analysis, Writing - review & editing. Pavel A. Levkin: Methodology, Funding acquisition, Supervision, Writing - review & editing.
Acknowledgments
Authors are grateful for support by the ERC Starting Grant (DropCellArray 337077), the ERC Proof-of-Concept is stated doubled Grant (CellPrintArray 768929), and the HGF-ERC-0016 Grant from the Helmholtz Associations´ Initiative and Networking Fund. Authors also would like to thank Karlsruhe Nano Micro Facility, a Helmholtz Research Infrastructure at Karlsruhe Institute of Technology, Sagi Eppel for programming of hydrogel detection software and Dr. Pascal Friedrich for valuable suggestions and discussions. Parts of this work were carried out with the support of the Karlsruhe Nano Micro Facility, a Helmholtz Research Infrastructure at Karlsruhe Institute of Technology.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2020.100053.
Appendix A. Supplementary data
The following is/are the Supplementary data to this article:
References
- 1.Allazetta S., Negro A., Lutolf M.P. Microfluidic programming of compositional hydrogel landscapes. Macromol. Rapid Commun. 2017;38(15):1700255. doi: 10.1002/marc.201700255. [DOI] [PubMed] [Google Scholar]
- 2.Neto A., Vasconcelos N., Oliveira S., Ruiz-Molina D., Mano J. High-throughput topographic, mechanical, and biological screening of multilayer films containing mussel-inspired biopolymers. Adv. Funct. Mater. 2016;26:2745–2755. [Google Scholar]
- 3.Anderson D.G., Levenberg S., Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 4.Tourniaire G., Collins J., Campbell S., Mizomoto H., Ogawa S., Thaburet J.-F., Bradley M. Polymer microarrays for cellular adhesion. Chem. Commun. 2006;20:2118–2120. doi: 10.1039/b602009g. [DOI] [PubMed] [Google Scholar]
- 5.Khan F., Smith J.O., Kanczler J.M., Tare R.S., Oreffo R.O.C., Bradley M. Discovery and evaluation of a functional ternary polymer blend for bone repair: translation from a microarray to a clinical model. Adv. Funct. Mater. 2013;23(22):2850–2862. [Google Scholar]
- 6.Santoro R., Venkateswaran S., Amadeo F., Zhang R., Brioschi M., Callanan A., Agrifoglio M., Banfi C., Bradley M., Pesce M. Acrylate-based materials for heart valve scaffold engineering. Biomater. Sci. 2018;6(1):154–167. doi: 10.1039/c7bm00854f. [DOI] [PubMed] [Google Scholar]
- 7.Wu M., Bridle H., Bradley M. Targeting Cryptosporidium parvum capture. Water Res. 2012;46(6):1715–1722. doi: 10.1016/j.watres.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Venkateswaran S., Wu M., Gwynne P.J., Hardman A., Lilienkampf A., Pernagallo S., Blakely G., Swann D.G., Gallagher M.P., Bradley M. Bacteria repelling poly(methylmethacrylate-co-dimethylacrylamide) coatings for biomedical devices. J. Mater. Chem. B. 2014;2(39):6723–6729. doi: 10.1039/c4tb01129e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel A.K., Tibbitt M.W., Celiz A.D., Davies M.C., Langer R., Denning C., Alexander M.R., Anderson D.G. High throughput screening for discovery of materials that control stem cell fate. Curr. Opin. Solid State Mater. Sci. 2016;20(4):202–211. [Google Scholar]
- 10.Hansen A., Mjoseng H.K., Zhang R., Kalloudis M., Koutsos V., de Sousa P.A., Bradley M. High-density polymer microarrays: identifying synthetic polymers that control human embryonic stem cell growth. Adv. Healthc. Mater. 2014;3(6):848–853. doi: 10.1002/adhm.201300489. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R., Mjoseng H.K., Hoeve M.A., Bauer N.G., Pells S., Besseling R., Velugotla S., Tourniaire G., Kishen R.E.B., Tsenkina Y., Armit C., Duffy C.R.E., Helfen M., Edenhofer F., de Sousa P.A., Bradley M. A thermoresponsive and chemically defined hydrogel for long-term culture of human embryonic stem cells. Nat. Commun. 2013;4:1335. doi: 10.1038/ncomms2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F., Mei Y., Langer R., Anderson D.G. High throughput optimization of stem cell microenvironments. Comb. Chem. High Throughput Screen. 2009;12(6):554–561. doi: 10.2174/138620709788681916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenfeld A., Levkin P.A. High-Throughput combinatorial synthesis of stimuli-responsive materials. Adv. Biosys. 2019;3:1800293. doi: 10.1002/adbi.201800293. [DOI] [PubMed] [Google Scholar]
- 14.Jia J., Coyle R.C., Richards D.J., Berry C.L., Barrs R.W., Biggs J., James Chou C., Trusk T.C., Mei Y. Development of peptide-functionalized synthetic hydrogel microarrays for stem cell and tissue engineering applications. Acta Biomater. 2016;45:110–120. doi: 10.1016/j.actbio.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R., Liberski A., Khan F., Diaz–Mochon J.J., Bradley M. Inkjet fabrication of hydrogel microarrays using in situ nanolitre-scale polymerisation. Chem. Commun. 2008;11:1317–1319. doi: 10.1039/b717932d. [DOI] [PubMed] [Google Scholar]
- 16.Zhang T., Wang Y., Zhang F., Chen X., Hu G., Meng J., Wang S. Bio-inspired superhydrophilic coatings with high anti-adhesion against mineral scales. NPG Asia Mater. 2018;10:e471. [Google Scholar]
- 17.Shi L., Liu X., Wang W., Jiang L., Wang S. A self-pumping dressing for draining excessive biofluid around wounds. Adv. Mater. 2019;31:1804187. doi: 10.1002/adma.201804187. [DOI] [PubMed] [Google Scholar]
- 18.Le N.N.T., Zorn S., Schmitt S.K., Gopalan P., Murphy W.L. Hydrogel arrays formed via differential wettability patterning enable combinatorial screening of stem cell behaviour. Acta Biomater. 2016;34:93–103. doi: 10.1016/j.actbio.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popova A.A., Schillo S.M., Demir K., Ueda E., Nesterov-Mueller A., Levkin P.A. Droplet-Array (DA) sandwich chip: a versatile platform for high-throughput cell screening based on superhydrophobic-superhydrophilic micropatterning. Adv. Mater. 2015;27(35):5217–5222. doi: 10.1002/adma.201502115. [DOI] [PubMed] [Google Scholar]
- 20.Neto A.I., Demir K., Popova A.A., Oliveira M.B., Mano J.F., Levkin P.A. Fabrication of hydrogel particles of defined shapes using superhydrophobic-hydrophilic micropatterns. Adv. Mater. 2016;28(35):7613–7619. doi: 10.1002/adma.201602350. [DOI] [PubMed] [Google Scholar]
- 21.Mason T.G., Weitz D.A. Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids. Phys. Rev. Lett. 1995;74(7):1250–1253. doi: 10.1103/PhysRevLett.74.1250. [DOI] [PubMed] [Google Scholar]
- 22.Mason T.G. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes–Einstein equation. Rheol. Acta. 2000;39(4):371–378. [Google Scholar]
- 23.Weeks E.R., Crocker J.C., Levitt A.C., Schofield A., Weitz D.A. Three-dimensional direct imaging of structural relaxation near the colloidal glass transition. Science. 2000;287(5453):627–631. doi: 10.1126/science.287.5453.627. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczyk A., Oelschlaeger C., Willenbacher N. Visualization of micro-scale inhomogeneities in acrylic thickener solutions: a multiple particle tracking study. Polymer. 2015;58:170–179. [Google Scholar]
- 25.Mason T.G., Ganesan K., van Zanten J.H., Wirtz D., Kuo S.C. Particle tracking microrheology of complex fluids. Phys. Rev. Lett. 1997;79(17):3282–3285. [Google Scholar]
- 26.Schober L., Büttner E., Laske C., Traube A., Brode T., Traube A.F., Bauernhansl T. Cell dispensing in low-volume range with the immediate drop-on-demand technology (I-DOT) J. Lab. Autom. 2015;20(2):154–163. doi: 10.1177/2211068214562450. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Scheiger J.M., Tronser T., Long C., Demir K., Wilson C.L., Kuzina M.A., Levkin P.A. Hydrogels with preprogrammable lifetime via UV-Induced polymerization and degradation. Adv. Funct. Mater. 2019;29(33):1902906. [Google Scholar]
- 28.Chen M., Zhong M., Johnson J.A. Light-controlled radical polymerization: mechanisms, methods, and applications. Chem. Rev. 2016;116(17):10167–10211. doi: 10.1021/acs.chemrev.5b00671. [DOI] [PubMed] [Google Scholar]
- 29.Chapman R., Gormley A.J., Stenzel M.H., Stevens M.M. Combinatorial low-volume synthesis of well-defined polymers by enzyme degassing. Angew. Chem. Int. Ed. 2016;55(14):4500–4503. doi: 10.1002/anie.201600112. [DOI] [PubMed] [Google Scholar]
- 30.Chapman R., Gormley A.J., Herpoldt K.-L., Stevens M.M. Highly controlled open vessel RAFT polymerizations by enzyme degassing. Macromolecules. 2014;47(24):8541–8547. [Google Scholar]
- 31.Oytun F., Kahveci M.U., Yagci Y. Sugar overcomes oxygen inhibition in photoinitiated free radical polymerization. J. Polym. Sci. A Chem. 2013;51(8):1685–1689. [Google Scholar]
- 32.Gormley A.J., Chapman R., Stevens M.M. Polymerization amplified detection for nanoparticle-based biosensing. Nano Lett. 2014;14(11):6368–6373. doi: 10.1021/nl502840h. [DOI] [PubMed] [Google Scholar]
- 33.Kröber P., Delaney J.T., Perelaer J., Schubert U.S. Reactive inkjet printing of polyurethanes. J. Mater. Chem. 2009;19(29):5234–5238. [Google Scholar]
- 34.Holmes P.F., Bohrer M., Kohn J. Exploration of polymethacrylate structure-property correlations: advances towards combinatorial and high-throughput methods for biomaterials discovery. Prog. Polym. Sci. 2008;33(8):787–796. doi: 10.1016/j.progpolymsci.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.