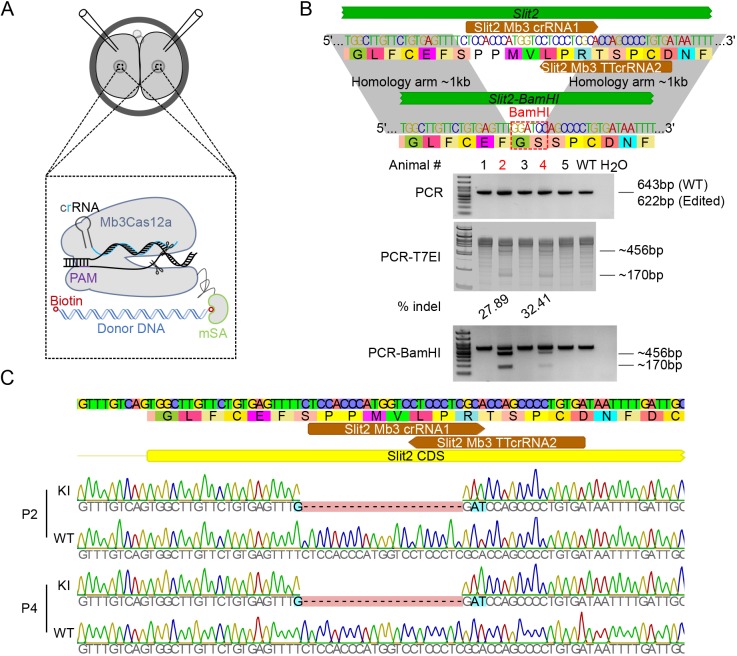
Fig. 3.
Generation of a KI in Slit2 locus using Mb3Cas12a-mSA in mouse two-cell embryos. (A) Schematics showing the strategy used to generate a KI in two-cell mouse embryos. Mb3Cas12a-mSA mRNA, crRNA and biotinylated donor DNA template are microinjected into two-cell mouse embryos. Purple and cyan lines indicate the PAM sequence and spacer, respectively. The blue double-stranded helix indicates the donor DNA with biotin (red open circle). (B) The efficiency of Mb3Cas12a-mSA-mediated KI at the Slit2 locus in mouse. One crRNA harboring two 23 nt spacers recognizing one TTTV PAM and one TTV PAM sequence was used to target the Slit2 locus. Upper panel, the strategy used to generate Slit2-BamHI KI. The colored characters represent the DNA sequence, and the black characters in colored background indicate corresponding amino acids. Spacers used are shown in brown. The edited BamHI site is shown in red dashed square. Lower panel, PCR, PCR-T7EI (T7 endonuclease I assay) and PCR-BamHI digestion results showing the KI efficiency. The edited founders are indicated in red. The expected band sizes and the percentage of indels are indicated on the right and bottom of the gels, respectively. (C) Sanger sequencing results of Mb3Cas12a-mSA-mediated KI in pups #2 (P2) and #4 (P4) in Slit2 locus. Spacers used are shown in brown.