Abstract
Introduction
Cunninghamella bertholletiae although rarely causing mucormycosis, is responsible for the highest mortality among mucormycetes. The diagnosis of mucormycosis is challenged by the absence of specific biomarkers. Herein, we report a fatal case of C. bertholletiae infection and detection of its DNA in the serum by polymerase chain reaction (PCR).
Presentation of case
A 23-year-old male with refractory osteosarcoma was admitted with multiple lung metastases. He was on oral voriconazole prophylaxis after pulmonary aspergillosis. He suffered from fever during temporary neutropenia following chemotherapy and showed several neurological and respiratory symptoms. Despite liposomal-amphotericin B administration, the symptoms rapidly progressed, and he died five days after the onset of neurological symptoms.
We retrospectively evaluated the filamentous fungus isolated after his death from gastric juices. Based on the sequence of the internal transcribed spacer (ITS) region we identified the fungal isolate as C. bertholletiae. A 146-bp portion of the D1/D2 region was quantified by quantitative-PCR using DNA extracted from the serum. C. bertholletiae DNA load in the serum was 18.0 copies/μL on the day of onset of neurological symptoms, with the highest (101.0 copies/μL) on the day of his death.
Discussion
Detection of circulating DNA of mucormycetes in the blood would greatly enhance the diagnosis of mucormycosis. Rapid diagnosis might alleviate mortality due to mucormycosis.
Conclusion
The present case-report suggests that the quantification of C. bertholletiae DNA in the serum could be useful for the diagnosis and evaluation of mucormycosis pathogenesis in patients.
Keywords: Cunninghamella bertholletiae, Mucormycosis, Zygomycosis, Circulating DNA, Internal transcribed spacer region
Introduction
The incidence of mucormycosis has been increasing in the last two decades among immunocompromised patients [1]. The average annual incidence rate of mucormycosis increases with age from 0.3/million in children aged 0–9 years to 3.9/million in patients aged > 89 years [1]. Patients with diabetes and malignancies, and those receiving immunosuppressive agents, deferoxamine therapy, or broad-spectrum antimicrobial drugs are at the highest risk of zygomycosis (mucormycosis) [2]. The overall mortality rate due to mucormycosis has been reported at almost 100 % in disseminated subtypes, 85 % in gastrointestinal subtypes, 76 % or more in pulmonary subtypes, 62 % in rhinocerebral subtypes, and 10 % in cutaneous subtypes [3]. Cunninghamella bertholletiae rarely causes mucormycosis, but it is responsible for the highest mortality among all mucormycetes. Despite its increasing frequency, mucormycosis remains challenging to diagnose because the fungi belonging to the order Mucorales are difficult to culture and have no specific biomarker. Zygomycetes have recently been detected by semi-nested polymerase chain reaction (PCR) targeting Mucorales 18S ribosomal DNA and sequencing of formalin-fixed paraffin-embedded biopsy samples [4]. However, obtaining biopsy samples from immunocompromised host might not be feasible. Recently, quantitative PCR (qPCR) has been used to detect circulating Mucor/Rhizopus, Lichtheimia, and Rhizomucor DNA in the serum [4].Herein, we report a fatal case of C. bertholletiae infection and the detection of its DNA in the serum by PCR, retrospectively.
Case report
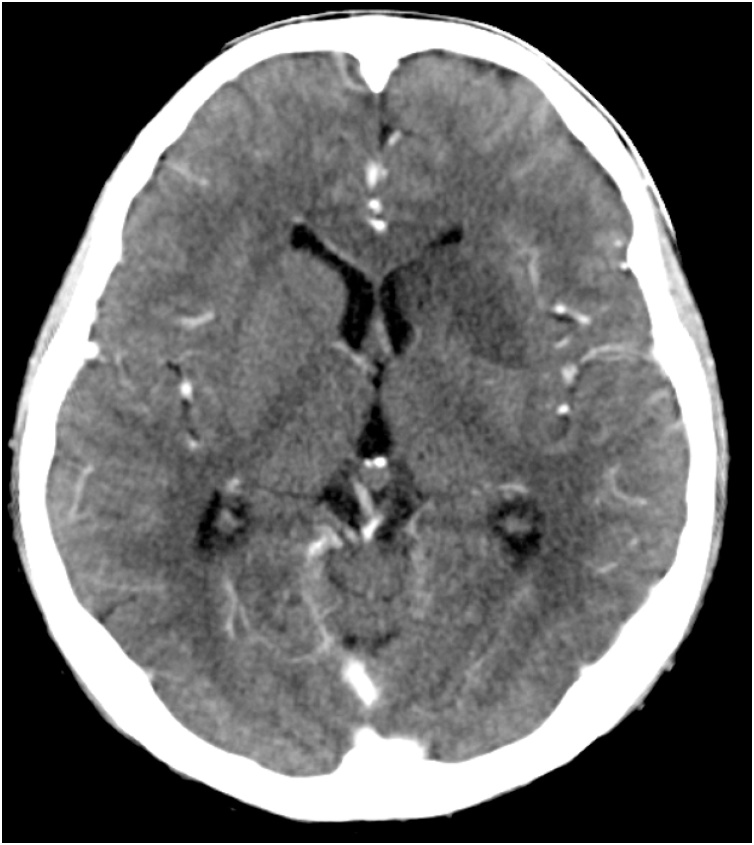
A 23-year-old male with refractory Stage IIb osteosarcoma of the left tibia was admitted to our pediatric unit with multiple lung metastases. He was initially diagnosed with osteosarcoma at 16 years of age, and this was his fourth relapse. He had been treated with multi-agent chemotherapy, but gradually became refractory to it. He was on oral voriconazole (VRCZ) prophylaxis after pulmonary aspergillosis. Six months after VRCZ prophylaxis, he had a fever during a period of neutropenia following chemotherapy. He suddenly showed several neurological and respiratory symptoms, including left ptosis, loss of vision, left pupillary dilatation, disorientation, and dyspnea. A brain computed tomography (CT) scan showed low-density areas in the left caudate nucleus, putamen, and frontal lobe, which were suggestive of cerebral infarction (Fig. 1). A chest CT scan showed consolidation with surrounding ground-glass opacity in the right upper lobe. Sputum, cerebrospinal fluid (CSF), and blood culture were negative. Serological examinations including assays for beta-D-glucan and antigens for aspergillosis, candidiasis, and cryptococcosis were negative. We suspected mucormycosis based on the neurological symptoms and administered liposomal-amphotericin B (L-AMB). However, the symptoms rapidly progressed, and he died five days after the onset of the neurological symptoms.
Fig. 1.
Brain computed tomography scan at the onset of neurological symptoms. It showed low-density areas in the left caudate nucleus, putamen, and frontal lobe.
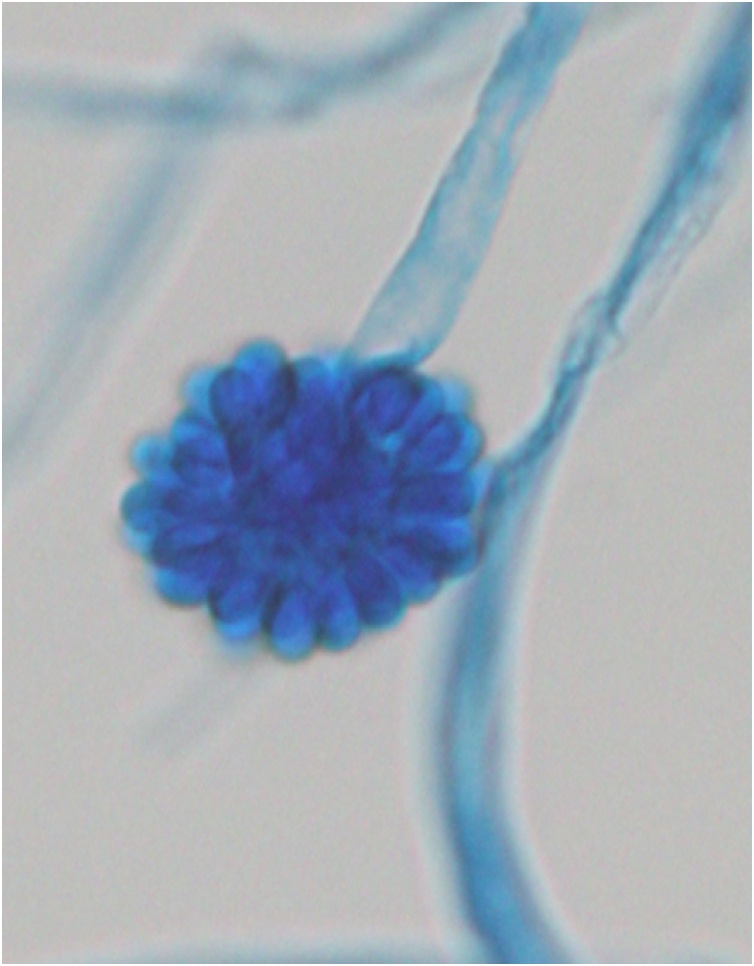
After his death, a filamentous fungus was isolated from gastric juice. The fungus was very fast growing on Sabouraud’s agar, and the colonies turned white to gray and powdery as they developed. Microscopic observations revealed simple sporangiophores forming a swollen, terminal vesicle around which single-celled, globose to ovoid sporangiola developed on swollen denticles (Fig. 2).
Fig. 2.
Cunninghamella bertholletiae isolated from gastric juice after the patient’s death. Lactophenol cotton blue staining. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
(×400) A sporangiophore terminates with a swollen vesicle, which is covered by single-spored sporangioles.
Materials and methods
Informed consent
Informed consent was obtained from the patient’s family and the healthy volunteers.
Identification of a filamentous fungus in gastric juice
DNA was extracted from the fungus using the DNeasy Plant Mini Kit (Qiagen K.K., Tokyo, Japan), according to the manufacturer’s instructions. The fungus was identified from its internal transcribed spacer (ITS) region sequence using DNA of an isolated fungus and the universal PCR primers for fungi (forward primer, 5′-GGAAGTAAAAGTCGTAACAAGG-3′; reverse primer 5′-TCCTCCGCTTATTGATATGC-3′), corresponding to a ∼500-bp amplicon.
Amplification and quantification of the serum fungal DNA
DNA was extracted from 200 μL each of the serum and cell-free CSF of the patient using the QIAamp DNA Blood Mini Kit (Qiagen), according to the manufacturer’s instructions. A 146-bp portion of the D1/D2 region was amplified by PCR using appropriate primers (forward primer, 5′-AGGCATAGCTTACCCGGATT-3′; reverse primer 5′-TCTTCGTTTCCAAGCCAAAC-3′). The amplicon was shorter than the one used for cultured fungus because the fungal DNA fragments in the blood are typically shorter than 200 bp. PCR was conducted on an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermal cycling conditions were as follows: an initial denaturation step at 95 °C for 20 s, followed by 40 cycles of 3 s at 95 °C and 30 s at 60 °C. The assay was designed against a four-point standard curve constructed using Cunninghamella-specific sequence-containing plasmids at concentrations ranging from 10 to 104 copies/μL. The threshold cycle value for each sample was plotted on the standard curve. The copy number of C. bertholletiae DNA was calculated and expressed as the copy number per microliter of serum.
Results
The fungus obtained from the culture of gastric juice sample was identified as C. bertholletiae by the sequence of the ITS region. A 146-bp portion of the D1/D2 region of the fungus was amplified by PCR using DNA extracted from gastric juice. By using the same primers, the D1/D2 region could be amplified and quantified by PCR using DNA extracted from the serum samples collected from the first day of the onset of neurological symptoms (day 1) to the day of his death (day 4) (Table 1A) although no bacteria or fungus could be cultured from the blood sample. Cunninghamella bertholletiae DNA load in the serum was 18.0 copies/μL on day 1, with the highest (101.0 copies/μL) on day 4. The specificity of amplification was confirmed by a dissociation curve analysis. C. bertholletiae DNA load in the serum was not detected approximately two and a half months before the development of the neurological symptoms (day -75). The D1/D2 sequence was undetectable in four healthy volunteers (Table 1B).
Table 1A.
Numbers of copies of D1/D2 sequence of Cunninghamella bertholletiae per microliter serum in the patient from the day of neurological symptom onset (day 1) until the day he died (day 4).
| Patient | ||||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | |
| Cunninghamella DNA copy number (copies/μL serum) | 18.0 | 15.9 | 9.9 | 101.0 |
Table 1B.
Numbers of copies of D1/D2 sequence of Cunninghamella bertholletiae per microliter serum in four healthy volunteers (Nos.1–4).
| Nos. 1–4:healthy volunteers | ||||
|---|---|---|---|---|
| No | 1 | 2 | 3 | 4 |
| Cunninghamella DNA copy number (copies/μL serum) | ND | ND | ND | ND |
ND: not detected.
Discussion
Species belonging to the genera Rhizopus, Mucor, and Lichtheimia account for 70 %–80 % of all cases of mucormycosis. In contrast, species of the genera Cunninghamella, Apophysomyces, Saksenaea, Rhizomucor, Cokeromyces, Actinomucor, and Syncephalastrum individually are responsible for less than 1 %–5 % of the reported cases of mucormycosis [5]. C. bertholletiae infections have been recently identified with increased frequency among immunocompromised hosts [6]. Mortality has been high (81 %) regardless of the underlying disease or infection site [7].
The route of infection in our patient was speculated to be via the airway or gastrointestinal tract. Although we isolated C. bertholletiae from gastric juice after his death, he did not present any gastrointestinal tract-related symptoms. We speculate that the patient developed pneumonia due to the breakthrough of C. bertholletiae through the airway while receiving VRCZ therapy and orally ingested the bronchial secretion containing pathogens. The infection progressively disseminated through the blood vessels, leading to cerebral infarction in the left putamen and caudate nucleus.
Currently, the diagnosis of mucormycosis is difficult because there is no specific biomarker for its diagnosis. It may not be feasible to obtain deep-tissue samples or bronchoalveolar lavage (BAL) from a patient with neutropenia and low platelet count. Kasai et al. developed a quantitative PCR assay for zygomycosis (mucormycosis) using the plasma, BAL, and lung tissue in a rabbit model of experimental pulmonary zygomycosis (mucormycosis) [8]. The assay was significantly more sensitive than conventional culture methods (100 % vs. 67 %, P < 0.05) [8].
The qPCR using primers and probes targeting the 18S rDNA genes of Mucor/Rhizopus, Lichtheimia, and Rhizomucor in immunocompromised patients can be used to detect DNA levels as low as a few femtograms [9]. High-risk patients of mucormycosis should be routinely examined for Mucor/Rhizopus, Lichtheimia, and Rhizomucor in that order because circulating DNA of the order Mucorales can be detected before the occurrence of the first clinical or radiological signs [9]. The minor mucormycosis-causing fungi such as C. bertholletiae might be difficult to detect in advance because screening for each species is expensive and time-consuming.
Development of a biomarker for mucormycosis in the blood would be highly beneficial for the diagnosis. Rapid diagnosis might alleviate mortality due to mucormycosis. The present findings suggest that the quantification of C. bertholletiae DNA in the serum is useful for the diagnosis and evaluation of pathogenesis of mucormycosis.
Ethical approval
All procedures performed in this study involving human participants were approved by the ethical review committee of the Kyoto Prefectural University of Medicine.
Funding
This work was supported by JSPS KAKENHI [grant number (A) 25253095].
Contributions
MM conceptualized and designed the study. RH and MM contributed to data collection. All authors contributed to the analysis and the interpretation of the data. RH and MM drafted the manuscript. All authors critically reviewed and approved the final version of the manuscript.
Declaration of Competing Interest
None.
Acknowledgments
We want to thank Editage (www.editage.jp) for English language editing.
References
- 1.Bitar D., Van Cauteren D., Lanternier F., Dannaoui E., Che D., Dromer F. Increasing incidence of zygomycosis (mucormycosis), France, 1997–2006. J Clin Microbiol. 2009;15:1395–1401. doi: 10.3201/eid1509.090334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez C.E., Rinaldi M.G., Sugar A.M. Zygomycosis. Infect Dis Clin North Am. 2002;16:895–914. doi: 10.1016/s0891-5520(02)00037-5. [DOI] [PubMed] [Google Scholar]
- 3.Roden M.M., Zaoutis T.E., Buchanan W.L., Knudsen T.A., Sarkisova T.A., Schaufele R.L. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis. 2005;41:634–653. doi: 10.1086/432579. [DOI] [PubMed] [Google Scholar]
- 4.Hammond S.P., Bialek R., Milner D.A., Petschnigg E.M., Baden L.R., Marty F.M. Molecular methods to improve diagnosis and identification of mucormycosis. J Clin Microbiol. 2011;49:2151–2153. doi: 10.1128/JCM.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes M.Z., Lewis R.E., Kontoyiannis D.P. Mucormycosis caused by unusual mucormycetes, non-Rhizopus, -Mucor, and -Lichtheimia species. Clin Microbiol Rev. 2011;24:411–445. doi: 10.1128/CMR.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellanger A.P., Berceanu A., Rocchi S., Valot B., Chauchet A., Belin N. Development of a quantitative PCR detecting Cunninghamella bertholletiae to help in diagnosing this rare and aggressive mucormycosis. Bone Marrow Transplant. 2018;53:1180–1183. doi: 10.1038/s41409-018-0194-5. [DOI] [PubMed] [Google Scholar]
- 7.Garey K.W., Pendiand S.L., Huynh V.T., Bunch T.H., Jensen G.M., Pursell K.J. Cunninghamella bertholletiae infection in a bone marrow transplant patient: amphotericin lung penetration, MIC determinations, and review of the literature. Pharmacotherapy. 2001;21:855–860. doi: 10.1592/phco.21.9.855.34560. [DOI] [PubMed] [Google Scholar]
- 8.Kasai M., Harrington S.M., Francesconl A., Petraitis V., Petraitiene R., Beveridge M.G. Detection of a molecular biomarker for zygomycetes by quantitative PCR assays of plasma, bronchoalveolar lavage, and lung tissue in a rabbit model of experimental pulmonary zygomycosis. J Clin Microbiol. 2008;36:3690–3702. doi: 10.1128/JCM.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millon L., Larosa F., Lepiller Q., Legrand F., Rocchi S., Daguindau E. Quantitative polymerase chain reaction detection of circulating DNA in serum for early diagnosis of mucormycosis in immunocompromised patients. Clin Infect Dis. 2013;56:e95–101. doi: 10.1093/cid/cit094. [DOI] [PubMed] [Google Scholar]