Figure 2: In silico structural modifications to gallinamide A and their resulting docking scores.
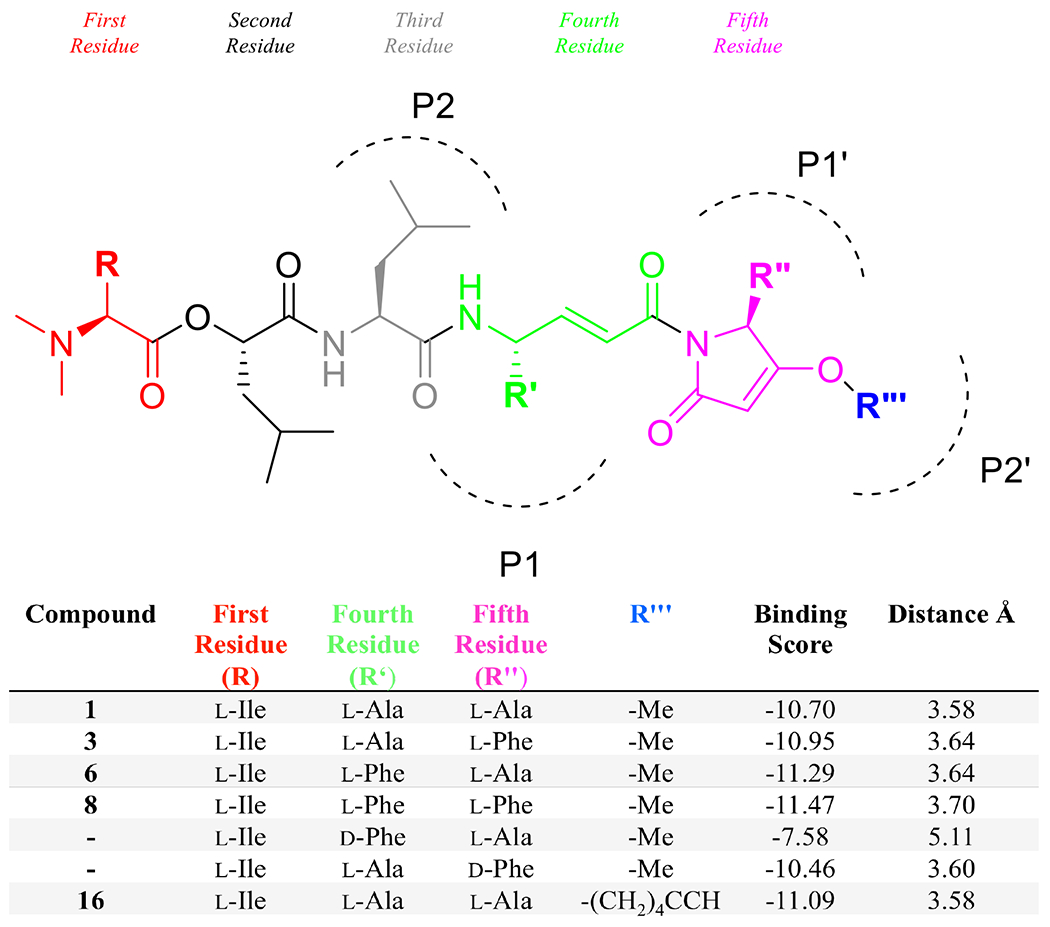
The residues within the gallinamide analogs are labeled according to Schechter-Berger nomenclature, with the fourth and fifth residues residing at P1 and P1’, respectively. Changes to the substrate residues (at R, R’, R” and R”’) are shown with a corresponding binding score, calculated with GBVI/WSA dG. Binding scores with a lower number (more negative) are predicted to bind with higher affinity.
