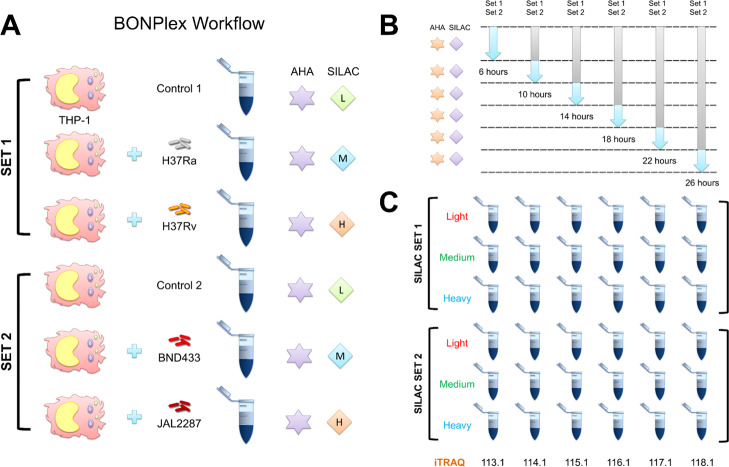
Figure 3.
Overview of the BONPlex workflow and labeling. (A) Workflow comprises selective enrichment of newly synthesized proteins with BONCAT (AHA) and pulsed SILAC labeling. Two sets of SILAC were used to label THP-1 cells infected with different mycobacterial strains. (B) SILAC and AHA labels were incorporated (after 1 h of methionine, lysine, and arginine depletion) in specific time windows postinfection. (C) Cells were harvested, proteins were digested, and time points were labeled with iTRAQ reagents as shown. The supernatant was selected for LC–MS/MS, and the 18-plexed data generated from each run was analyzed using the HyperQuant pipeline.