Abstract
Triclosan (TCS) has been a widely used antibacterial agent in medical and personal care products in the last few decades. Methyl TCS (MTCS) is the major biotransformation product of TCS through replacement of the hydroxyl group with methoxy. Previous studies revealed that MTCS showed reduced toxicity but enhanced environmental persistence, when compared with TCS. Till date, the toxicological molecular mechanisms of TCS and MTCS remain to be clarified. This study aimed to investigate the transcriptomic changes in HepG2 cells induced by TCS and MTCS using microarray chips and to identify key target genes and related signal pathways. The microarray data showed that there were 1664 and 7144 differentially expressed genes (DEGs) in TCS- and MTCS-treated groups, respectively. Gene ontology (GO) enrichment and Kyoto Encyclopedia of genes and genomes (KEGG) analysis revealed that TCS and MTCS induced overlapping as well as distinct transcriptome signatures in HepG2 cells. Both TCS and MTCS could result in various biological responses in HepG2 cells mainly responding to biosynthetic and metabolic processes but probably through different regulatory pathways. Among the selected 50 GO terms, 9 GO terms belonging to the cellular component category were only enriched in the MTCS group, which are mainly participating in the regulation of cellular organelle’s function. KEGG analysis showed that 19 and 59 pathway terms were separately enriched in TCS and MTCS groups, with only seven identical pathways. The selected 10 TCS-specific signal pathways are mainly involved in cell proliferation and apoptosis, while the selected 10 MTCS-specific pathways mainly take part in the regulation of protein synthesis and modification. The overall data suggested that MTCS induced more enriched DEGs, GO terms, and pathway terms than TCS. In conclusion, compared with TCS, MTCS presents lower polarity and stronger lipophilicity, enabling MTCS to cause more extensive transcriptomic changes in HepG2 cells, activate differentiated signal pathways, and finally lead to differences in biological responses.
Introduction
Triclosan (TCS) is seriously affecting people’s lives through a variety of ways.1 First, TCS is widely used as an antibacterial agent in various products, including hand sanitizer, soap, and toothpaste. Approximately 75% of commercial soaps were added with TCS or its derivatives, according to the statistics of the US Food and Drug Administration.2 Second, TCS added in food packaging could possibly migrate to food and then enter into human bodies.3 Furthermore, TCS can be applied in medical products such as TCS-coated antibacterial sutures to reduce surgical-site infections4 and TCS-contained composite materials to treat periodontitis.5 Methyl TCS (MTCS) is a dominant biotransformation product of TCS under aerobic conditions. The substitution of the hydroxyl group by methoxy (Supporting Information, Figure S1) makes MTCS lose its antibacterial property but acquire stronger lipophilicity, bioaccumulation, and environmental persistence, as compared with TCS.6
With the extensive use of TCS-containing products, TCS and MTCS have been detected in various environmental and biological media such as water bodies, sediments, animals, and even human bodies.1,7 Wei et al. found that the concentration ranges of TCS and MTCS in tributary of Yangtze River in Nanjing of China reached 247–433 and 403–453 ng/L, respectively.8 TCS in urine samples collected from Chinese children ranged from none-detected to 681.38 μg/L, and TCS in breast milk samples from Spanish ranged from 0.25 to 2.1 μg/L.9,10 Koppen et al. reported that urinary excretion of TCS was 2.72 μg/L in mother and 1.23 μg/L in child of Belgian.11 The concentrations of TCS and MTCS were found to be 0.126–0.161 μg/L in blood samples and 0.211–0.254 μg/L in urine samples collected in Wenzhou, China.12
TCS is currently recognized as an environmental endocrine disruptor, which can interfere with series of physiological functions of reproductive, immune, nervous, and endocrine systems. As the major biotransformation product of TCS, previous studies revealed that MTCS showed reduced toxicity but enhanced environmental persistence, as compared with TCS. Stenzel et al. found that TCS (with environmentally relevant concentrations of 0.4–40 μg/L) could have adverse effects on metamorphosis, fecundity, and fertility in adult zebrafish and delay the maturation of offsprings.13 The sublethal concentration of TCS (0.37 mg/L) had been proved to cause DNA strand break and have genotoxicity in Indian carp Labeo rohita.14 TCS and MTCS at environmental concentrations significantly influenced the growth and reproductive performance of earthworm Eisenia andrei.15 Gaume et al. reported both TCS and MTCS caused toxic effects at micromolar levels in immune cells, as evidenced by changes in the morphology and density of hemocytes.16 Macedo et al. also found that TCS and MTCS could impact embryonic development of Danio rerio and Paracentrotus lividus.17 Moreover, TCS has been identified as a potential carcinogen by US Environmental Protection Agency. TCS exposure experiments in mice revealed that TCS was mainly accumulated in liver,18 finally resulting in liver dysfunction, fatty liver, fibrosis, and even hepatic cancer.19−21
The toxicological mechanism of TCS and MTCS has not yet been well elucidated till date. As far as known, TCS functions as a mitochondrial uncoupler to disrupt adenosine 5′-triphosphate (ATP) generation and inhibit degranulation in mammal cells.22 TCS exposure in L02 normal cells induced upregulation of purine and amino acid metabolism, accumulation of lipid, and interference of energy metabolism in vitro.23 Our previous work also investigated the toxic effects of TCS and MTCS in human HepG2 cells, showing that TCS displayed a higher cytotoxicity than MTCS through different molecular pathways.24 In human body, formation of the MTCS–human serum albumin (HSA) complex has been detected, which could affect protein and endocrine functions,24 but the potential toxicology mechanism of MTCS is still very limited. Changes in RNA transcripts are the first cellular response linking genomes to biological outcomes. Thus, investigation on the transcriptomic changes under TCS and MTCS exposure may be helpful in explaining the molecular basis of the toxicity and facilitate in-depth toxicology research.
Genome-wide transcriptomics have advantages of high sensitivity, high throughput, high speed, and high integration, which are being used to comprehensively analyze the transcriptomic changes and reveal the molecular mechanism caused by TCS. Transcriptomics researches in zebrafish revealed the role of the liver as a target organ for TCS toxicity, and the liver steatosis resulted from increased fatty acid synthetase and uptake and suppression of β-oxidation.25,26 In embryonic liver of chicken, TCS could induce xenobiotic metabolism and activate the thyroid hormone receptor-mediated downstream signaling.27 Furthermore, a newly developed human-reduced transcriptomics approach is presently used to qualitatively and quantitatively assess the profiles of AHR-regulated genes and pathways in HepG2 cells exposed to TCS.28 In this study, we analyzed the widespread transcriptomic changes induced by TCS and MTCS to differentiate their toxic effects and better understand the underline toxicological mechanisms. Enrichment analysis of biological functions and signal pathways were performed to screen the target/key genes and molecular pathways distinctly or overlappingly responded to TCS and MTCS. Our results may help to provide new insights into further toxicological studies of TCS and MTCS.
Results and Discussion
Overall Transcriptomic Changes Induced by TCS and MTCS
Till date, studies on the toxicity and mechanism of TCS and MTCS are not sufficient. Our previous studies suggested that TCS showed stronger cytotoxicity than MTCS in HepG2 cells, in terms of cell proliferation inhibition, DNA damages, cell cycle arrest, and apoptosis. Although both TCS and MTCS (over 10 μM) induced oxidative DNA damages and initiated DNA damage repair processes, they blocked cell cycle progress at different stages through differential modulation on cyclin A2 and CDK2 genes.24 Moreover, TCS activated the p53-mediated apoptotic pathway in a caspase-independent manner, while MTCS induced apoptosis dependent on caspases.24 These findings indicated that replacement of hydrogen with methoxy in TCS not only changed the physicochemical properties but also altered its interactions with cellular biomolecules, which may ultimately result in significant differences in biological outcomes of TCS and MTCS.
The statistics and fold change (FC) of the differentially expressed genes (DEGs) in TCS- and MTCS-treated groups are listed in Table 1, and the scatter plot of TCS or MTCS versus control group are shown in the Supporting Information (Figure S2). As compared with the control group, there were 1664 DEGs in the TCS-treated group, of which 946 genes (56.9%) were upregulated and 718 genes (43.1%) were downregulated. Although MTCS seems to be more inert in terms of cytotoxicity in HepG2 cells compared to TCS,24 MTCS induced more extensive transcriptomic changes with a total of 7144 DEGs, among which 2261 genes (31.6%) were upregulated and 4883 genes (68.3%) were downregulated. As compared with TCS, the higher lipophilicity of MTCS possibly makes it easier to bind with cellular biomolecules6 and trigger broader cellular responses in vitro. These transcription expression changes could further be used to screen potential molecular targets involved in differential biological responses induced by MTCS and TCS.
Table 1. Screening Results of DEGs in HepG2 Cellsa.
| groups | number of DEGs | upregulated genes | downregulated genes |
|---|---|---|---|
| TCS vs control | 1664 | 946 | 718 |
| MTCS vs control | 7144 | 2261 | 4883 |
Note: The screening criteria was set as the absolute FC ≥ 2 and p < 0.05. The FD referred to the ratio of gene expression levels in TCS or MTCS group to that in control group.
In this study, real time-quantitative polymerase chain reaction (RT-qPCR) assay was applied to verify the reliability of the microarray method, by randomly selecting two genes with opposite expression changes. As shown in Table 2, as compared to the control group, results of the microarray chip assay revealed that the gene of B-cell lymphoma/leukemia 2 (bcl2) was upregulated to 2.83 and 2.36 times in TCS and MTCS-treated groups, respectively; and proto-oncogene mdm2 was downregulated to 0.45 and 0.51 times, respectively. As shown in Table 2, the results of RT-qPCR were consistent with that of the microarray chip assay. Slight differences may be because of the different sensitivity of the two detecting methods.
Table 2. Expression Levels of bcl2 and mdm2 Measured with Microarray and RT-qPCR.
| microarray |
RT-qPCR |
|||
|---|---|---|---|---|
| gene | TCS | MTCS | TCS | MTCS |
| Bcl2 | 2.83 (up) | 2.36 (up) | 1.58 (up) | 1.95 (up) |
| Mdm2 | 0.45 (down) | 0.51 (down) | 0.83 (down) | 0.81 (down) |
HepG2 cells were treated with 20 μM of TCS or MTCS for 6 h, and related gene transcription expressions were measured with the microarray assay or RT-qPCR assay (gapdh was the internal reference).
Interestingly, there were hundreds of DEGs that showed completely opposite changes under treatment of TCS and MTCS in this study. For example, expression of the serine/threonine kinase 4 (stk4) was upregulated to 3.50 times of the control group in TCS-treated samples, while its expression in MTCS was downregulated to 0.49 times. Similarly, expression of the atm gene (ataxia telangiectasia mutated) was also increased in TCS but decreased in MTCS. ATM plays a central role in repairing DNA double-strand breaks, as well as in cell cycle arrest and apoptosis to maintain genomic stability. Overexpression of ATM may indicate a cellular response to DNA damages caused by TCS (but not MTCS) because of its mitochondrial uncoupler capability. The oppositely expressed genes between TCS and MTCS may be helpful in explaining the molecular basis of their different biological effects and be used in in-depth mechanism research.
Gene Ontology Enrichment Analysis
Gene ontology (GO) enrichment analysis is a widely used comprehensive resource for computational analysis on biological function of genes.29 Usually, GO information consists of three main categories: cellular component (CC), molecular function (MF), and biological process (BP). Through GO enrichment analysis based on DEGs induced by TCS and MTCS, we aimed to screen targeted key GO terms and analyze the inner relationships between the target GO terms, finally to establish a complex structural network system.
As shown in the Supporting Information (Data Set Sheet 1), GO enrichment analysis showed that there were 59 GO terms enriched in the TCS treatment group, including 8 for MF, 5 for CC, and 46 for BP. Consistent with the trend of transcription changes, MTCS treatment showed significantly more enrichment of 514 GO terms, including 70 for MF, 97 for CC, and 347 for BP (Supporting Information, Data Set Sheet 2). The broad grouping of GO terms into the three main categories is shown in Figure 1. The GO terms were ranked according to the number of enriched genes, and the first 50 GO terms (Supporting Information, Data Set Sheet 3 and Sheet 4) were selected for further comparative analysis between TCS and MTCS.
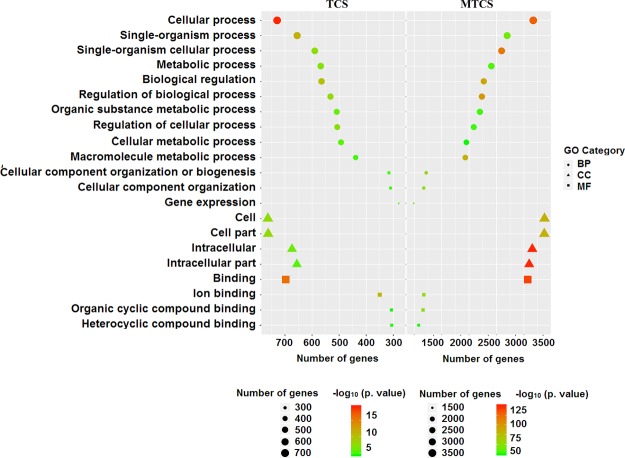
Figure 1.
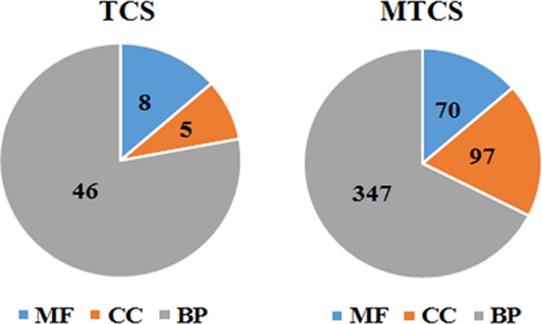
Broad grouping of GO terms into three main groups: BP, CC, and MF.
Comparative Analysis of GO Terms Induced by TCS and MTCS Treatment
Further comparison of the selected 50 GO terms induced by TCS and MTCS was conducted to investigate their distinct as well as overlapping biological mechanisms. There were 21 identical GO terms, both contained by two treatments (Figure 2 and Supporting Information, Data Set Sheet 5), of which 4 GO terms were associated with MF, 4 with CC, and 13 with BP. Briefly, GO: 0097159 (organic cyclic compound binding) is responsible for selectively interacting and noncovalently binding with an organic cyclic compound. GO: 1901363 (heterocyclic compound binding) is in charge of selectively interacting with an heterocyclic compound. In addition, some metabolism-related GO terms such as GO: 0071704 (an organic substance metabolic process) and its subclass GO: 0043170 (a macromolecule metabolic process) were also enriched in TCS and MTCS groups. These two GO terms participate in chemical reactions and metabolic pathways of organic substances and macromolecules, respectively. Taken together, the comprehensive classification of GO terms showed that both TCS- and MTCS-treated HepG2 cells mainly responded to biosynthetic processes and metabolic pathways. The results of Ajao et al. also showed that TCS can upshift the rate of glucose consumption in mammalian cells.30 The latest data of Zhang suggested that TCS could promote the progression of hepatocellular carcinoma by accelerating energy metabolism.23 Both TCS and MTCS could impair energy metabolism and amino acid synthesis in developing zebrafish embryos.31 These results were consistent with the GO enrichment analysis in this study.
Figure 2.
Overlapping GO terms induced by TCS and MTCS treatment. The HepG2 cells were treated with TCS and MTCS at 20 μM for 6 h, RNA was extracted and measured with the Affymetrix Human U133 plus 2.0 chip. The enriched GO terms classified as BP, CC, and MF were represented with circle, triangle, and square, respectively. The enrichment degree of GO terms was sorted based on numbers of enriched DEGs and p values and were indicated by different colors in each category.
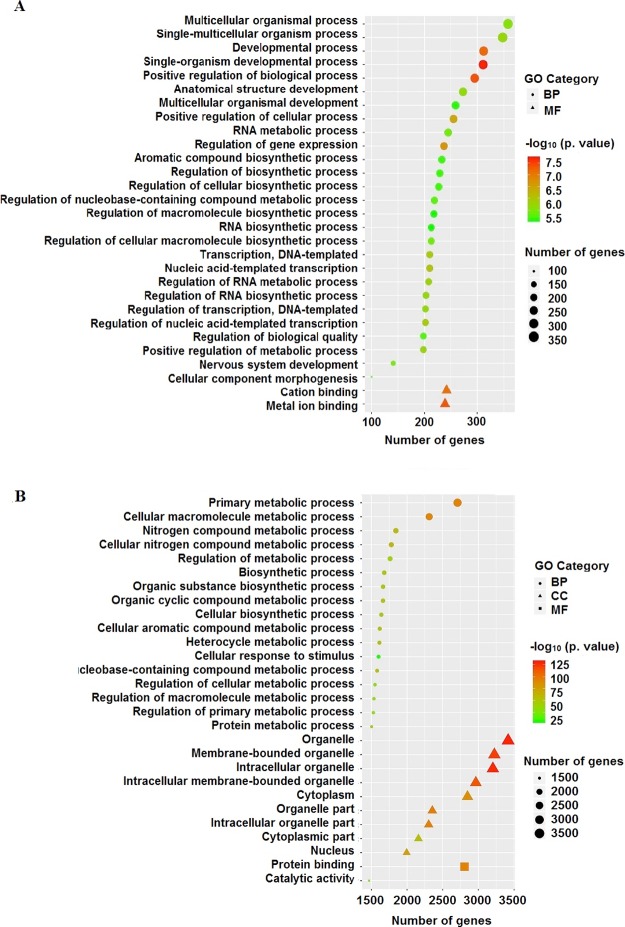
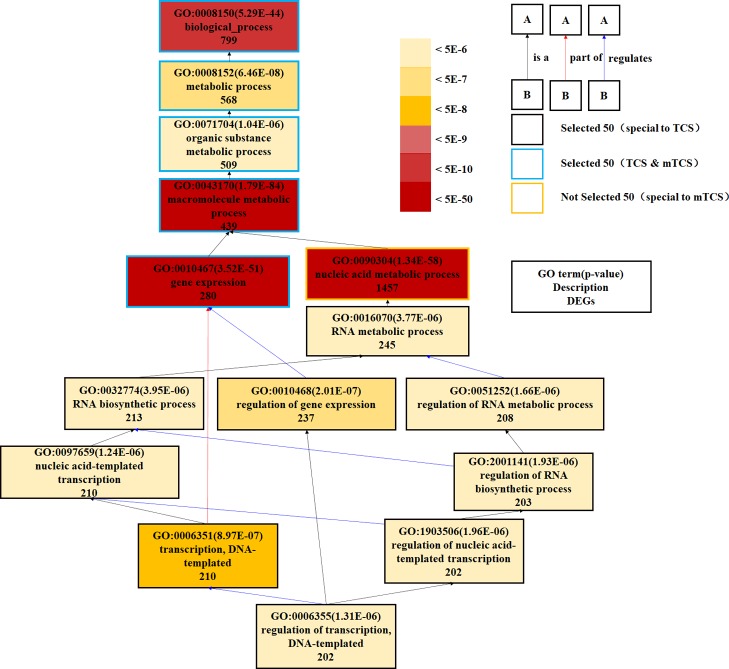
The 29 specific GO terms responded to TCS other than MTCS and are listed in Figure 3A (Supporting Information, Data Set Sheet 6), including 2 for MF and 27 for BP category. The two MF GO term, GO: 0043169 (cation binding) and GO: 0046872 (metal ion binding) play roles in selectively noncovalent interaction with cation or metal ions in cells. The other 27 BP GO terms mainly participate in transcriptional biosynthesis, metabolic regulation, and developmental processes. A partial directed acyclic graph (DAG) based on TCS individual GO terms is shown in Figure 4, providing a rough interrelation network among the metabolically related GO terms. The GO: 0016070, GO: 0032774, GO: 0010468, GO: 0051252, GO: 0097659, GO: 2001141, GO: 1903506, GO: 0006351, and GO: 0006355 terms form a cross network to regulate the biosynthesis process and metabolic process of macromolecule. This finding was consistent with the results of Affymetrix miRNA 4.0 microarrays on male zebrafish brain exposed to TCS, which also indicated that TCS-changed miRNAs significantly influenced translation, transcription, DNA-templated, and protein transportation.32 Our study further proved that TCS exposure could enhance glycolysis metabolism through accelerating glucose decomposition and ATP production.33
Figure 3.
Specific GO terms induced by TCS or MTCS treatment. The HepG2 cells were treated with TCS (A) and MTCS (B) at 20 μM for 6 h, RNA was extracted and measured with Affymetrix Human U133 plus 2.0 chip. GO enrichment belonging to BP, CC, and MF were represented in circle, triangle, and square, respectively. The enrichment degree of GO terms was sorted based on numbers of enriched DEGs and p values and were indicated by different colors.
Figure 4.
DAG for partial enriched GO terms specifically responded to TCS treatment. Different colors of rectangles represent different enrichment degree of GO terms based on the responded DEG numbers and p values. Different colors of lines with arrows refer to the relationship between two GO terms: the black means B is a A; the red means B is part of A; and the blue means B regulates A.
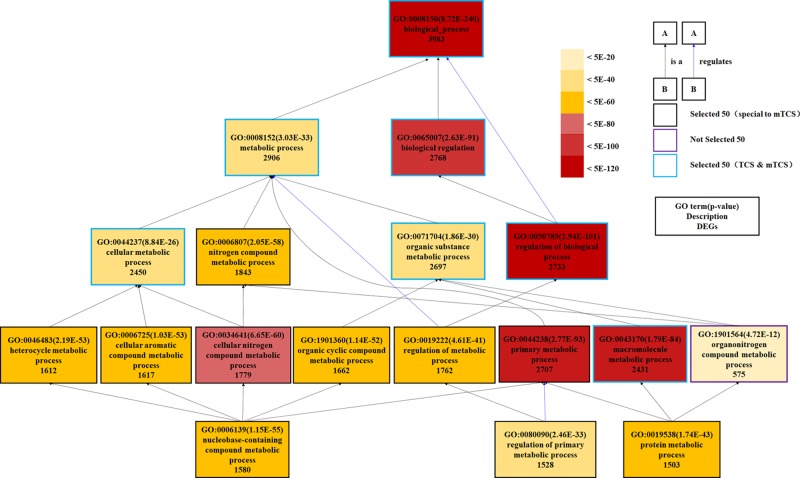
Figure 3B (Supporting Information, Data Set Sheet 7) summarized the 29 GO terms that specifically responded to MTCS treatment, including 2 for MF, 9 for CC, and 17 for BP category. Most of the MTCS-responded GO terms under BP category were mainly associated with metabolic regulation (13/17) on chemicals, including nitrogen compound, organic cyclic compound, aromatic compound, macromolecule, and proteins. Fu et al. also reported that MTCS could induce changes in the metabolic pathways of starch, sucrose, and nitrogen, as well as changes in the biosynthetic pathways of fatty acid, phenylalanine, tyrosine, and tryptophan in zebrafish embryos.34 The DAG graph, as shown in Figure 5, displayed the hierarchical or containment relationship among the 10 GO terms involved in the metabolic processes of nitrogen compound and organic substance (including GO: 0006807, GO: 0046483, GO: 0006725, GO: 0034641, GO: 1901360, GO: 0019222, GO: 0044238, GO: 0006139, GO: 0080090, and GO: 0019538). Expectedly, among the selected 50 GO terms, there were 9 GO terms under the CC category that specifically responded to MTCS but not to TCS. From GO: 0043226, GO: 0043229, GO: 0043227, GO: 0043231 to GO: 0005634, a cascade subclass relationship can be established. These data remind us that MTCS could regulate the biological functions of membrane-enclosed organelle, including nucleus and mitochondria.
Figure 5.
DAG for partial enriched GO terms of BP category specifically responded to MTCS treatment. Different colors of rectangles represent different enrichment degree of GO terms based on DEG numbers and p values. The lines with arrows refer to the relationship between two GO terms: the black color means B is a A; the red means B is part of A; and the blue means B regulates A.
Taken together, in terms of biological functions, TCS and MTCS can influence intracellular biosynthesis and metabolism after a short period of exposure through differentiated regulation pathways. In addition, MTCS can specifically regulate the function of CC organelle such as membrane-enclosed nucleus, which might also result from its higher lipophilicity with cellular membrane components, when compared with TCS.
Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
Kyoto Encyclopedia of genes and genomes (KEGG) is a bioinformatics database for systematic analysis of genomic information, including cellular biochemical processes, pathway networks, and enzymatic reactions. KEGG enrichment analysis based on DEGs is helpful in revealing the signal transduction pathways and the underlying regulatory mechanism.35
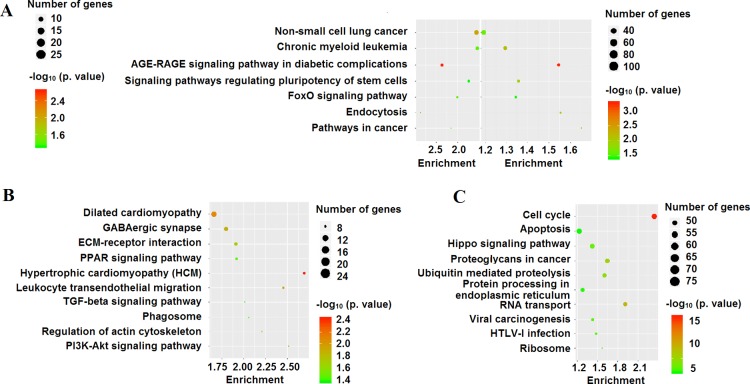
In this study, we further performed KEGG enrichment analysis to explore associated signal pathways induced by TCS and MTCS in HepG2 cells. Identification of signal pathways would improve the understanding on the function of key genes and shed light on related molecular mechanisms. Under the criterion of p < 0.05, 19 pathway terms were enriched after TCS treatment (Supporting Information, Data Set Sheet 8) and 59 pathway terms for MTCS (Supporting Information, Data Set Sheet 9). The homologous comparison and screening analysis revealed that there were seven overlapping pathways responding to both TCS and MTCS treatment (Figure 6A, Supporting Information, Data Set Sheet 10). The other 12 and 52 pathways separately responded to TCS and MTCS treatment and are listed in detail in the Supporting Information (Data Set Sheet 11 and Sheet 12). Figure 6B,C shows the selected 10 KEGG pathways, according to the number of enriched genes.
Figure 6.
KEGG enrichment analysis in HepG2 cells under the influence of TCS and MTCS. The HepG2 cells were treated with TCS and MTCS at 20 μM for 6 h, RNA was extracted and measured with Affymetrix Human U133 plus 2.0 chip. (A) Overlapping KEGG pathways induced by TCS and MTCS treatment. (B) Selected 10 specific KEGG pathways induced by TCS treatment. (C) Selected 10 specific KEGG pathways induced by MTCS treatment.
Comparative Analysis of Signal Pathways Induced by TCS and MTCS Treatment
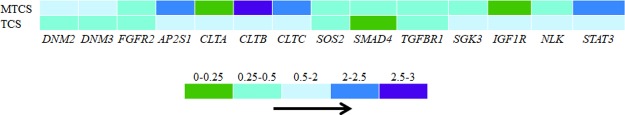
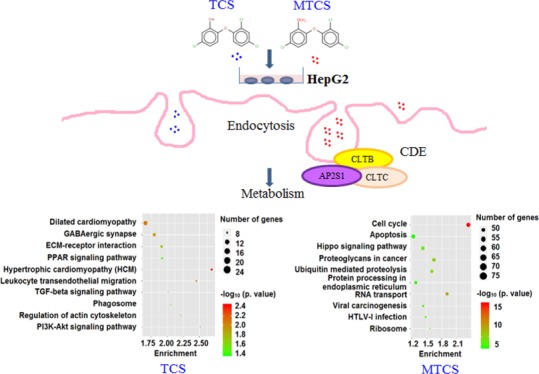
As shown in Figure 6A and Supporting Information (Data Set Sheet 10), the pathways in cancer (hsa05200) enriched a maximum number of DEGs induced by TCS and MTCS treatment and followed by pathways of endocytosis (hsa04144) and forkhead box O (FoxO) signaling (hsa04068). There were 18 and 79 enriched DEGs for TCS and MTCS, respectively, in the endocytosis (hsa04144) pathway, which is an important mechanism in interacting with the environment and ingesting extracellular materials through clathrin-dependent (CDE) or clathrin-independent ways.36 Extracellular compounds could transport into cells through the CDE pathway and bind to the adaptor-related protein complexes, enabling their rapid removal from plasma membrane.36 The FoxO signaling pathway (hsa04068) is involved in various physiological and pathological events, including cell proliferation, apoptosis, cell cycle control, glucose metabolism, oxidative stress resistance, and longevity.37 Yamaguchi et al. found that activation of the FoxO signaling pathway and the altered expressions of downstream target genes played an important role in cell cycle arrest and growth suppression in liver cancer cells.38 According to the pathway map of hsa04068 from KEGG (https://www.kegg.jp/kegg-bin/show_pathway?hsa04068), as a central hub, the activity of FoxO is regulated by a series of genes. The expressions of negative regulators such as SOS2, SMAD4, and TGFBR1 were significantly downregulated after both TCS and MTCS treatment (Figure 7).
Figure 7.
Expression levels of related genes involved in endocytosis (hsa04144) and FoxO signaling pathway (hsa04068). Different colors represent the fold intervals of transcription expression in TCS/MTCS groups, as compared to the control group.
Although TCS and MTCS seemed to have overlapping functions in inducing the hsa05200, hsa04144, and hsa04068 signaling pathways, there were differences in molecular regulation. For the hsa05200 pathway, MTCS enriched far more genes (111 DEGs) as compared to TCS (28 DEGs), suggesting that the carcinogenic molecular pathways of MTCS is far more complicated than TCS. Furthermore, some signal transduction pathways interacting with hsa05200 such as basal transcription factors (hsa03022) specifically responded to MTCS.39 Notably, expressions of some negative regulators of FoxO, including SGK3, IGF1R, and NLK were only inhibited by MTCS but not by TCS. The expressions of the positive FoxO regulator STAT3 and regulators involved in endocytosis such as the clathrin light chain B, C (CLTB and CLTC) and AP 2 subunit sigma 1 (AP2S1) were also specifically upregulated by MTCS treatment (Figure 7), indicating that CDE endocytosis and differential FoxO signal pathways play an important role in MTCS-induced BP.
The specific signal pathways for TCS treatment (Figure 6B, Supporting Information, Data Set Sheet 11) mainly regulate the cell proliferation and apoptosis, including phosphatidylinositol 3 kinase (PI3K)-protein kinase B (Akt) (hsa04151), transforming growth factor-β (TGF-β) (hsa04350), peroxisome proliferator-activated receptors (PPARs) signaling pathway (hsa03320), and extracellular matrix–receptor interaction (hsa04512). The PI3K/Akt signaling pathway is involved in regulating the basic cellular functions such as transcription, translation, proliferation, and survival. The Akt activity modulates multiple BP through phosphorylating substrates involved in apoptosis, protein synthesis, metabolism, and cell cycle.40 Activation of the PI3K/Akt pathway mediated by oncogene G-protein signaling modulator 2 (GPSM2) in HepG2 cells can subsequently promote cell proliferation, cell cycle progression, metastasis, and apoptosis inhibition.41 Conversely, inhibition of the PI3K/Akt signaling pathway could result in reduced mitochondrial membrane potential, elevated cell cycle arrest at G2/M transition and G1 phase, and increased mitochondrial pathway apoptosis in HepG2 cells.42 Our recent study also proved that TCS can cause oxidative damages, induce S phase cell cycle arrest,24 and promote glycolysis in HepG2 cells via the PI3K/Akt/FoxO pathway.33 In addition, other TCS-specific signal pathways found in this study could regulate the cell morphology, including actin cytoskeleton (hsa04810) and phagosome (hsa04145), and transendothelial migration of leukocyte for immune responses (hsa04670). These results suggested that TCS can quickly activate immune response and regulate cell proliferation and apoptosis in HepG2 cells.
Among the selected 10 MTCS-specific pathways (Figure 6C, Supporting Information, Data Set Sheet 12), four pathways participate in the regulation of protein synthesis and modification, including RNA transport (hsa03013), ribosome (hsa03010), protein processing in endoplasmic reticulum (hsa04141), and ubiquitin-mediated proteolysis (hsa04120). These results were well consistent with GO enrichment results that MTCS could quickly influence the functions of nucleus and mitochondria, thus affecting the formation and stability of genetic materials. In the remaining six pathways, the hippo signaling pathway (hsa04390) plays a role in regulating cell proliferation and differentiation through two genes of mammalian ste20-like kinase 1/2 (MST1/2) and cofactor large tumor suppressor.43 As the downstream transcriptional coactivators of the hippo pathway, the yes-related protein (YAP) and PDZ-binding site (TAZ) play a critical role in occurrence and development of primary liver cancer.44 Activation of the hippo/YAP signaling pathway can significantly inhibit HepG2 cell growth and induce apoptosis.45 In addition, the hippo/YAP signaling pathway could affect multisignaling pathways such as TGF-β and WNT/β-catenin and activate apoptotic pathways through interactions of YAP/TAZ with other transcription factors or signaling molecules.43,46
Conclusions
In conclusion, a short-term exposure of TCS and MTCS can induce comprehensive responses on cellular metabolism and biosynthesis in HepG2 cells. As compared with TCS, more GO terms were enriched after MTCS treatment, most of which participate in regulation of organelles functions. Both TCS and MTCS could induce alterations of pathways in cancer, endocytosis, and FoxO signaling pathways, but the changes in related signaling molecules were differentiated. Among the selected 10 MTCS-specific pathway terms, the hippo signaling pathway is responsible for regulating cell proliferation and differentiation. The TCS-specific PI3K-Akt, TGF-beta, and PPAR signaling pathways are important in regulation of cell proliferation and apoptosis. However, this study is only an investigative research on transcriptomic changes in TCS and MTCS using a selected dose comparatively higher than the environmentally relevant doses. Future work will focus on dose-dependent transcriptomics study in the ranges of lower concentrations. Moreover, because transcriptomic changes only indicate potential upcoming biological events, cross-analysis of transcriptomics with other omics such as metabolomic and proteomic is essential for in-depth investigation to exploit the toxicology mechanism of TCS and MTCS.
Materials and Methods
Materials
Cell Line and Media
HepG2 were purchased from American Type Culture Collection and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (Invitrogen, Paisley, UK) in a cell culture incubator at 37 °C under 5% CO2, as described previously.24 HepG2 cells were cultured and continuously passaged in culture dishes. Exposing concentrations of TCS and MTCS (CAS: 3380-34-5 and CAS: 4640-01-1, Dr. Ehrensorfer, Germany, purity >97%) in this study were chosen, according to our previous results.24 TCS and MTCS stock solutions (20 mM) were prepared using dimethyl sulfoxide (DMSO, Sigma, MO, USA) and then were diluted with DMEM. The HepG2 cells were treated with 20 μM of TCS/MTCS for 6 h maintaining the concentration of DMSO at 0.1% (v/v). The control HepG2 cells were treated with DMSO (0.1%, v/v). The treatment conditions used in this study were chosen based on our preliminary experiments, environmental concentrations of TCS/MTCS, and published papers.24,33,47 Previous results found that 20 μM of TCS/MTCS could cause moderate cytotoxicity in HepG2 cells after a short-term exposure.24 Exposure of TCS/MTCS (6 h) could result in extensive transcriptomic responses in HepG2 cells.33
Methodology
RNA Extraction and Purification
Total RNA samples were extracted using the TRIZOL agent (Life technologies, Carlsbad, CA, USA) from HepG2 cells after being exposed to 20 μM of TCS/MTCS or 0.1% of DMSO for 6 h. Total RNA was analyzed and qualified using the Agilent Bioanalyzer 2100 (Agilent technology, Santa Clara, CA, USA) through electrophoresis. The qualified RNA samples were purified by the RNeasy mini kit and RNase-Free DNase Set (QIAGEN, GmBH, Germany), according to the manufacturer’s instructions. The concentrations, integrity, and purity of the RNA samples were analyzed by a NanoDrop ND-2000 spectrophotometer. The results of RNA quality inspection are shown in the Supporting Information (Table S1).
Microarray Chip Assay
Gene expression profiling analysis was performed using the Affymetrix Human U133 plus 2.0 chips, which contained 47,000 transcripts and 38,500 human genes, covering almost all of the identified human genes. This microarray has been widely used to investigate the characteristics of DEGs in various physiological and pathological processes.48,49 The method of chip microarray analysis has been described in detail in the previous article.50,51
In brief, the biotin-labeled cRNA samples were prepared with a GeneChip 3′ IVT PLUS reagent kit (Affymetrix, Santa Clara, CA, USA) through amplification, labeling, and purification following the manufacturer’s instructions. The cRNA probes were orderly arranged on the chip vector and specifically hybridized with corresponding gene sequences in the microarray, according to the standard procedure of GeneChip hybridization, wash and stain kit (Affymetrix, Santa Clara, CA, USA). Then, the gene chip was scanned using a GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA, USA), and the fluorescence-converted raw data were read with a Command Console Software 4.0 (Affymetrix, Santa Clara, CA, USA).
According to the relative expression level of the treatment and control groups, DEGs were selected and divided into upregulated and downregulated groups. FC ≥ 2 or ≤ 0.5 was used as the screening criteria for DEGs. Then, the clusterProfiler package software from R/Bioconductor was used for bioinformatics analysis, including GO functional annotation and enrichment, and KEGG enrichment was used to explore the potential molecular mechanism of TCS or MTCS in HepG2 cells. Fisher’s exact test was applied for statistical analysis with screening criteria of p adjust < 0.05. STRING database was used to construct a potential gene interaction network.
RT-qPCR
Total RNA samples were extracted using the TRIZOL agent, as described above. Then, the Master Mixfan reverse transcription system was prepared with a RT-qPCR kit to reversely transcribe the total RNA into cDNA. The reverse transcription PCR reaction was performed at 37 °C for 15 min, followed with 95 °C for 10 min.
The 20 μL PCR reaction system consisted of 10 μL of a SYBR green agent, 1 μL of upstream and downstream primer mixture, and 9 μL of cDNA template. DNA amplification was carried out by the PCR reaction with three parallel samples. The amplification procedure was started at 95 °C for 10 min to denature the cDNA template and activate the Taq polymerase; and followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 63 °C for 30 s. The relative expression levels of target genes were calculated based on cycle threshold (Ct) values using the following formula: ΔCt = Ct (target gene) – Ct (gapdh); ΔΔCt = ΔCt(treated) – ΔCt(control); induction fold = 2(−ΔΔCt). The sequences of primer pairs for target genes are listed in Table 3.
Table 3. Sequence of Primers.
| gene | forward primer (FP) (5–3′) | reverse primer (RP) (5–3′) |
|---|---|---|
| Bcl2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| Mdm2 | GAATCATCGGACTCAGGTACATC | TCTGTCTCACTAATTGCTCTCCT |
| Gapdh | CCATGGAGAAGGCTGGGG | CAAAGTTGTCATGGATGACC |
Statistical Analysis
Data were expressed as mean ± standard error and were analyzed using the Student t-test with Bonferroni multiple test calibration to minimize the false positive selection. The significance thresholds were considered to be statistically significant (p < 0.05) and highly significant (p < 0.01).
Acknowledgments
The authors would like to thank the National Natural Science Foundation of China (nos. 41877371 and 41977366); the cooperative research program of Institute of Nature and Environmental Technology, Kanazawa University (19005); and the Joint Research Program of National Natural Science Foundation of China; and the Israel Science Foundation (no. 41561144007) for funding this study.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00075.
Chemical structure of TCS and MTCS and quality parameters of RNA (PDF)
GO terms enriched with TCS or MTCS treatment, selected 50 GO terms enriched with TCS or MTCS treatment, identical or specific GO terms induced by TCS or MTCS, KEGG pathways enriched with TCS or MTCS treatment, and the identical or specific KEGG pathways induced by TCS or MTCS (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Guerra P.; Teslic S.; Shah A.; Albert A.; Gewurtz S. B.; Smyth S. A. Occurrence and removal of triclosan in Canadian wastewater systems. Environ. Sci. Pollut. Res. Int. 2019, 26, 31873–31886. 10.1007/s11356-019-06338-w. [DOI] [PubMed] [Google Scholar]
- Bergstrom K. G. Update on antibacterial soaps: the FDA takes a second look at triclosans. J. Drugs Dermatol. 2014, 13, 501–503. [PubMed] [Google Scholar]
- Ruszkiewicz J. A.; Li S.; Rodriguez M. B.; Aschner M. Is Triclosan a neurotoxic agent?. J. Toxicol. Environ. Health B Crit. Rev. 2017, 20, 104–117. 10.1080/10937404.2017.1281181. [DOI] [PubMed] [Google Scholar]
- Ahmed I.; Boulton A. J.; Rizvi S.; Carlos W.; Dickenson E.; Smith N.; Reed M. The use of triclosan-coated sutures to prevent surgical site infections: a systematic review and meta-analysis of the literature. BMJ Open 2019, 9, e029727 10.1136/bmjopen-2019-029727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminu N.; Chan S.-Y.; Yam M.-F.; Toh S.-M. A dual-action chitosan-based nanogel system of triclosan and flurbiprofen for localised treatment of periodontitis. Int. J. Pharm. 2019, 570, 118659. 10.1016/j.ijpharm.2019.118659. [DOI] [PubMed] [Google Scholar]
- Clayborn A. B.; Toofan S. N.; Champlin F. R. Influence of methylation on the antibacterial properties of triclosan in Pasteurella multocida and Pseudomonas aeruginosa variant strains. J. Hosp. Infect. 2011, 77, 129–133. 10.1016/j.jhin.2010.09.021. [DOI] [PubMed] [Google Scholar]
- Yeh A.; Marcinek D. J.; Meador J. P.; Gallagher E. P. Effect of contaminants of emerging concern on liver mitochondrial function in Chinook salmon. Aquat. Toxicol. 2017, 190, 21–31. 10.1016/j.aquatox.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.; Yang X.; Watson P.; Yang F.; Liu H. A cyclodextrin polymer membrane-based passive sampler for measuring triclocarban, triclosan and methyl triclosan in rivers. Sci. Total Environ. 2019, 648, 109–115. 10.1016/j.scitotenv.2018.08.151. [DOI] [PubMed] [Google Scholar]
- Azzouz A.; Rascón A. J.; Ballesteros E. Simultaneous determination of parabens, alkylphenols, phenylphenols, bisphenol A and triclosan in human urine, blood and breast milk by continuous solid-phase extraction and gas chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2016, 119, 16–26. 10.1016/j.jpba.2015.11.024. [DOI] [PubMed] [Google Scholar]
- Li X.; Ying G.-G.; Zhao J.-L.; Chen Z.-F.; Lai H.-J.; Su H.-C. 4-Nonylphenol, bisphenol-A and triclosan levels in human urine of children and students in China, and the effects of drinking these bottled materials on the levels. Environ. Int. 2013, 52, 81–86. 10.1016/j.envint.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Koppen G.; Govarts E.; Vanermen G.; Voorspoels S.; Govindan M.; Dewolf M.-C.; Den Hond E.; Biot P.; Casteleyn L.; Kolossa-Gehring M.; Schwedler G.; Angerer J.; Koch H. M.; Schindler B. K.; Castaño A.; López M. E.; Sepai O.; Exley K.; Bloemen L.; Knudsen L. E.; Joas R.; Joas A.; Schoeters G.; Covaci A. Mothers and children are related, even in exposure to chemicals present in common consumer products. Environ. Res. 2019, 175, 297–307. 10.1016/j.envres.2019.05.023. [DOI] [PubMed] [Google Scholar]
- Wang X.; Gao M.; Gao J.; Wang X.; Ma M.; Wang H. Extraction of triclosan and methyltriclosan in human fluids by in situ ionic liquid morphologic transformation. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018, 1092, 19–28. 10.1016/j.jchromb.2018.05.038. [DOI] [PubMed] [Google Scholar]
- Stenzel A.; Wirt H.; Patten A.; Theodore B.; King-Heiden T. Larval exposure to environmentally relevant concentrations of triclosan impairs metamorphosis and reproductive fitness in zebrafish. Reprod. Toxicol. 2019, 87, 79–86. 10.1016/j.reprotox.2019.05.055. [DOI] [PubMed] [Google Scholar]
- Hemalatha D.; Nataraj B.; Rangasamy B.; Shobana C.; Ramesh M. DNA damage and physiological responses in an Indian major carp Labeo rohita exposed to an antimicrobial agent triclosan. Fish Physiol. Biochem. 2019, 45, 1463–1484. 10.1007/s10695-019-00661-2. [DOI] [PubMed] [Google Scholar]
- Chevillot F.; Guyot M.; Desrosiers M.; Cadoret N.; Veilleux É.; Cabana H.; Bellenger J.-P. Accumulation and sublethal effects of triclosan and its transformation product methyl-triclosan in the earthworm Eisenia andrei exposed to environmental concentrations in an artificial soil. Environ. Toxicol. Chem. 2018, 37, 1940–1948. 10.1002/etc.4156. [DOI] [PubMed] [Google Scholar]
- Gaume B.; Bourgougnon N.; Auzoux-Bordenave S.; Roig B.; Le Bot B.; Bedoux G. In vitro effects of triclosan and methyl-triclosan on the marine gastropod Haliotis tuberculata. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012, 156, 87–94. 10.1016/j.cbpc.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Macedo S.; Torres T.; Santos M. M. Methyl-triclosan and triclosan impact embryonic development of Danio rerio and Paracentrotus lividus. Ecotoxicology 2017, 26, 482–489. 10.1007/s10646-017-1778-3. [DOI] [PubMed] [Google Scholar]
- Escarrone A. L. V.; Caldas S. S.; Primel E. G.; Martins S. E.; Nery L. E. M. Uptake, tissue distribution and depuration of triclosan in the guppy Poecilia vivipara acclimated to freshwater. Sci. Total Environ. 2016, 560-561, 218–224. 10.1016/j.scitotenv.2016.04.039. [DOI] [PubMed] [Google Scholar]
- Rodricks J. V.; Swenberg J. A.; Borzelleca J. F.; Maronpot R. R.; Shipp A. M. Triclosan: a critical review of the experimental data and development of margins of safety for consumer products. Crit. Rev. Toxicol. 2010, 40, 422–484. 10.3109/10408441003667514. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Vanlandingham M.; Wu Y.; Beland F. A.; Olson G. R.; Fang J.-L. Role of peroxisome proliferator-activated receptor alpha (PPARalpha) and PPARalpha-mediated species differences in triclosan-induced liver toxicity. Arch. Toxicol. 2018, 92, 3391–3402. 10.1007/s00204-018-2308-7. [DOI] [PubMed] [Google Scholar]
- Yueh M.-F.; Taniguchi K.; Chen S.; Evans R. M.; Hammock B. D.; Karin M.; Tukey R. H. The commonly used antimicrobial additive triclosan is a liver tumor promoter. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 17200–17205. 10.1073/pnas.1419119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherly L. M.; Shim J.; Hashmi H. N.; Kennedy R. H.; Hess S. T.; Gosse J. A. Antimicrobial agent triclosan is a proton ionophore uncoupler of mitochondria in living rat and human mast cells and in primary human keratinocytes. J. Appl. Toxicol. 2016, 36, 777–789. 10.1002/jat.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Shao X.; Zhao H.; Li X.; Wei J.; Yang C.; Cai Z. Integration of metabolomics and lipidomics reveals metabolic mechanisms of triclosan-induced toxicity in human hepatocytes. Environ. Sci. Technol. 2019, 53, 5406–5415. 10.1021/acs.est.8b07281. [DOI] [PubMed] [Google Scholar]
- Wang L.; Mao B.; He H.; Shang Y.; Zhong Y.; Yu Z.; Yang Y.; Li H.; An J. Comparison of hepatotoxicity and mechanisms induced by triclosan (TCS) and methyl-triclosan (MTCS) in human liver hepatocellular HepG2 cells. Toxicol. Res. 2019, 8, 38–45. 10.1039/c8tx00199e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggard D. E.; Noyes P. D.; Waters K. M.; Tanguay R. L. Phenotypically anchored transcriptome profiling of developmental exposure to the antimicrobial agent, triclosan, reveals hepatotoxicity in embryonic zebrafish. Toxicol. Appl. Pharmacol. 2016, 308, 32–45. 10.1016/j.taap.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Ling Y.; Jiang J.; Wang D.; Wang J.; Li J.; Wang X.; Wang H. Differential mechanisms regarding triclosan vs. bisphenol A and fluorene-9-bisphenol induced zebrafish lipid-metabolism disorders by RNA-Seq. Chemosphere 2020, 251, 126318. 10.1016/j.chemosphere.2020.126318. [DOI] [PubMed] [Google Scholar]
- Guo J.; Ito S.; Nguyen H. T.; Yamamoto K.; Tanoue R.; Kunisue T.; Iwata H. Effects of prenatal exposure to triclosan on the liver transcriptome in chicken embryos. Toxicol. Appl. Pharmacol. 2018, 347, 23–32. 10.1016/j.taap.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Xia P.; Zhang H.; Peng Y.; Shi W.; Zhang X. Pathway-based assessment of single chemicals and mixtures by a high-throughput transcriptomics approach. Environ. Int. 2020, 136, 105455. 10.1016/j.envint.2019.105455. [DOI] [PubMed] [Google Scholar]
- Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajao C.; Andersson M. A.; Teplova V. V.; Nagy S.; Gahmberg C. G.; Andersson L. C.; Hautaniemi M.; Kakasi B.; Roivainen M.; Salkinoja-Salonen M. Mitochondrial toxicity of triclosan on mammalian cells. oxicol. Rep. 2015, 2, 624–637. 10.1016/j.toxrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J.; Tan Y. X. R.; Gong Z.; Bae S. The toxic effect of triclosan and methyl-triclosan on biological pathways revealed by metabolomics and gene expression in zebrafish embryos. Ecotoxicol. Environ. Saf. 2019, 189, 110039. 10.1016/j.ecoenv.2019.110039. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu F.; Chen W. Exposure to triclosan changes the expression of microRNA in male juvenile zebrafish (Danio rerio). Chemosphere 2019, 214, 651–658. 10.1016/j.chemosphere.2018.09.163. [DOI] [PubMed] [Google Scholar]
- An J.; He H.; Yao W.; Shang Y.; Jiang Y.; Yu Z. PI3K/Akt/FoxO pathway mediates glycolytic metabolism in HepG2 cells exposed to triclosan (TCS). Environ. Int. 2020, 136, 105428. 10.1016/j.envint.2019.105428. [DOI] [PubMed] [Google Scholar]
- Fu J.; Gong Z.; Kelly B. C. Metabolomic profiling of zebrafish (Danio rerio) embryos exposed to the antibacterial agent triclosan. Environ. Toxicol. Chem. 2019, 38, 240–249. 10.1002/etc.4292. [DOI] [PubMed] [Google Scholar]
- Chen L.; Zhang Y.-H.; Wang S.; Zhang Y.; Huang T.; Cai Y.-D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PloS One 2017, 12, e0184129 10.1371/journal.pone.0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G. J.; McMahon H. T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Farhan M.; Wang H.; Gaur U.; Little P. J.; Xu J.; Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 2017, 13, 815–827. 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi F.; Hirata Y.; Akram H.; Kamitori K.; Dong Y.; Sui L.; Tokuda M. FOXO/TXNIP pathway is involved in the suppression of hepatocellular carcinoma growth by glutamate antagonist MK-801. BMC Canc. 2013, 13, 468. 10.1186/1471-2407-13-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Zheng R.; Zhang H.; He T. Pathway crosstalk analysis of microarray gene expression profile in human hepatocellular carcinoma. Pathol. Oncol. Res. 2015, 21, 563–569. 10.1007/s12253-014-9855-x. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Gong W.; Liang P.; Huang X.; You N.; Han K. Q.; Li Y. M.; Li J. Effects of AFP-activated PI3K/Akt signaling pathway on cell proliferation of liver cancer. Tumour Boil 2014, 35, 4095–4099. 10.1007/s13277-013-1535-z. [DOI] [PubMed] [Google Scholar]
- He X. Q.; Zhang Y. F.; Yu J. J.; Gan Y. Y.; Han N. N.; Zhang M. X.; Ge W.; Deng J. J.; Zheng Y. F.; Xu X. M. High expression of G-protein signaling modulator 2 in hepatocellular carcinoma facilitates tumor growth and metastasis by activating the PI3K/AKT signaling pathway. Tumour Boil 2017, 39, 1010428317695971. 10.1177/1010428317695971. [DOI] [PubMed] [Google Scholar]
- Li D.; Wei X.; Ma M.; Jia H.; Zhang Y.; Kang W.; Wang T.; Shi X. FFJ-3 inhibits PKM2 protein expression via the PI3K/Akt signaling pathway and activates the mitochondrial apoptosis signaling pathway in human cancer cells. Oncol. Lett. 2017, 13, 2607–2614. 10.3892/ol.2017.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauviel A.; Nallet-Staub F.; Varelas X. Integrating developmental signals: a Hippo in the (path)way. Oncogene 2012, 31, 1743–1756. 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- Valero V. 3rd; Pawlik T. M.; Anders R. A. Emerging role of Hpo signaling and YAP in hepatocellular carcinoma. J. Hepatocell. Carcinoma 2015, 2, 69–78. 10.2147/JHC.S48505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Wang H.; Wang L.; Tan R.; Zhu M.; Zhong X.; Zhang Y.; Chen B.; Wang L. Decursin inhibits the growth of HepG2 hepatocellular carcinoma cells via Hippo/YAP signaling pathway. Phytother Res. 2018, 32, 2456–2465. 10.1002/ptr.6184. [DOI] [PubMed] [Google Scholar]
- Van Haele M.; Moya I. M.; Karaman R.; Rens G.; Snoeck J.; Govaere O.; Nevens F.; Verslype C.; Topal B.; Monbaliu D.; Halder G.; Roskams T. YAP and TAZ heterogeneity in primary liver cancer: an analysis of its prognostic and diagnostic role. Int. J. Mol. Sci. 2019, 20, E638 10.3390/ijms20030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura J.; Węgrzyn P.; Zarębski A.; Władyka B.; Koj A. Identification of changes in the transcriptome profile of human hepatoma HepG2 cells stimulated with interleukin-1 beta. Biochim. Biophys. Acta 2004, 1689, 120–133. 10.1016/j.bbadis.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Boeckmans J.; Buyl K.; Natale A.; Vandenbempt V.; Branson S.; De Boe V.; Rogiers V.; De Kock J.; Rodrigues R. M.; Vanhaecke T. Transcriptomics data of a human in vitro model of non-alcoholic steatohepatitis exposed to elafibranor. Data Brief 2019, 25, 104093. 10.1016/j.dib.2019.104093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvengadam S. S.; OʼMalley M.; LaGuardia L.; Lopez R.; Wang Z.; Shadrach B. L.; Chen Y.; Li C.; Veigl M. L.; Barnholtz-Sloan J. S.; Pai R. K.; Church J. M.; Kalady M. F.; Walsh R. M.; Burke C. A. Gene expression changes accompanying the duodenal adenoma-carcinoma sequence in familial adenomatous polyposis. Clin. Transl. Gastroenterol. 2019, 10, e00053 10.14309/ctg.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J.; An J.; Zhang D.; Shang Y.; Zheng K.; Yang Y. Transcriptomic analyses of the biological effects of black carbon exposure to A549 cells. J. Environ. Manage. 2019, 246, 289–298. 10.1016/j.jenvman.2019.05.123. [DOI] [PubMed] [Google Scholar]
- An J.; Zhou Q.; Qian G.; Wang T.; Wu M.; Zhu T.; Qiu X.; Shang Y.; Shang J. Comparison of gene expression profiles induced by fresh or ozone-oxidized black carbon particles in A549 cells. Chemosphere 2017, 180, 212–220. 10.1016/j.chemosphere.2017.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.