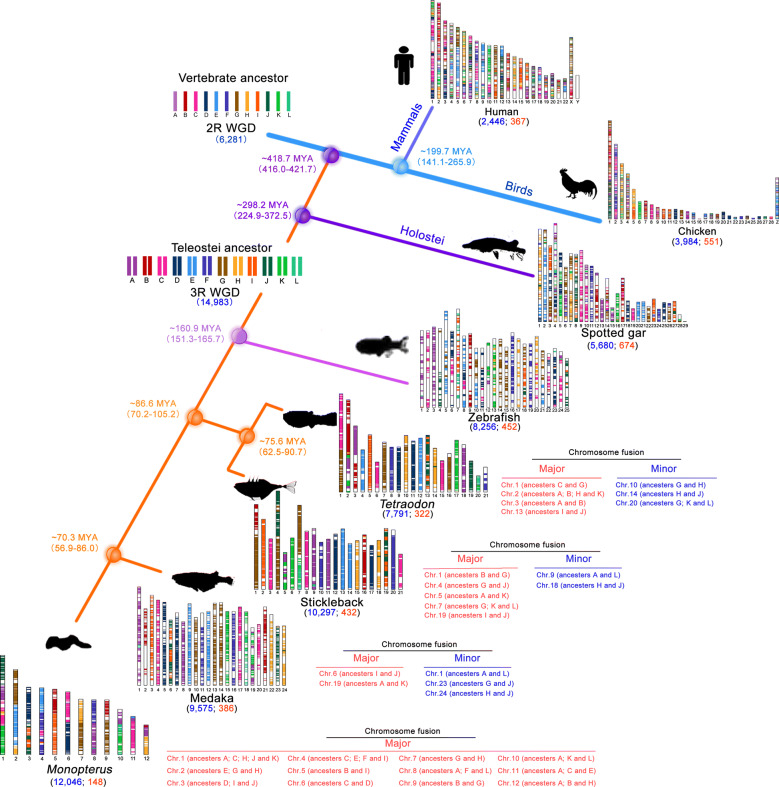
Fig. 1.
Phylogenomic trajectories of extant teleost fishes at chromosomal levels and chromosomal fusion events. Vertebrate genomes evolved from 12 ancestral chromosomes through chromosomal loss, fission and fusion, and syntenic block recombination, in addition to whole-genome duplications. The figure depicts the distribution of conserved syntenic blocks of chromosomes in teleosts (Monopterus albus, medaka, stickleback, Tetraodon and zebrafish), Holostei (spotted gar), birds (chicken) and mammals (human). Genomic blocks in each species originating from the ancestral chromosomes are shown in the same color. An additional genome duplication (the 3R WGD) took place in the teleost lineage after divergence from the Holostei (spotted gar), approximately 298 MYA, while the second round of genome duplication (the 2R WGD) occurred in the common ancestor of mammals, birds, Holostei and teleosts, approximately 500 MYA. Major and minor events of chromosome fusion in each species are shown on the right panels. Average divergence times are shown on the branch nodes. The number of conserved genes is shown in blue, and the number of conserved blocks is in orange