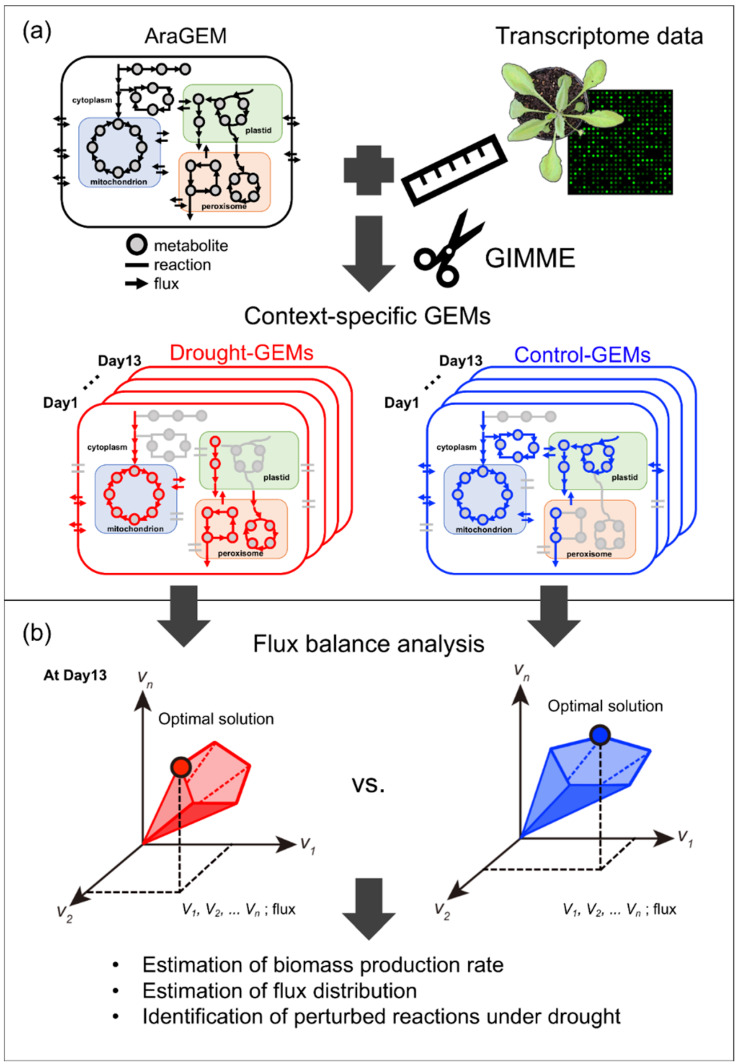
Figure 1.
Outline of this study for extracting information for the metabolic network by reconstructing context-specific genome-scale metabolic models (GEMs): (a) Transcriptome data were used to tailor the global GEM of A. thaliana and to obtain context-specific GEMs under progressive drought treatment (days 1–13). (b) Using flux balance analysis (FBA), the biomass production rate and flux distribution were estimated. Comparative analysis of the flux distribution between drought and control conditions was performed to identify candidate reactions associated with adaptation to drought. Drought and control GEMs are colored in red and blue, respectively. The eliminated reactions and metabolites are shown in grey. Examples of optimal solutions for flux distribution that lies on the border of the feasible space are represented by red and blue dots for drought and control GEMs, respectively.