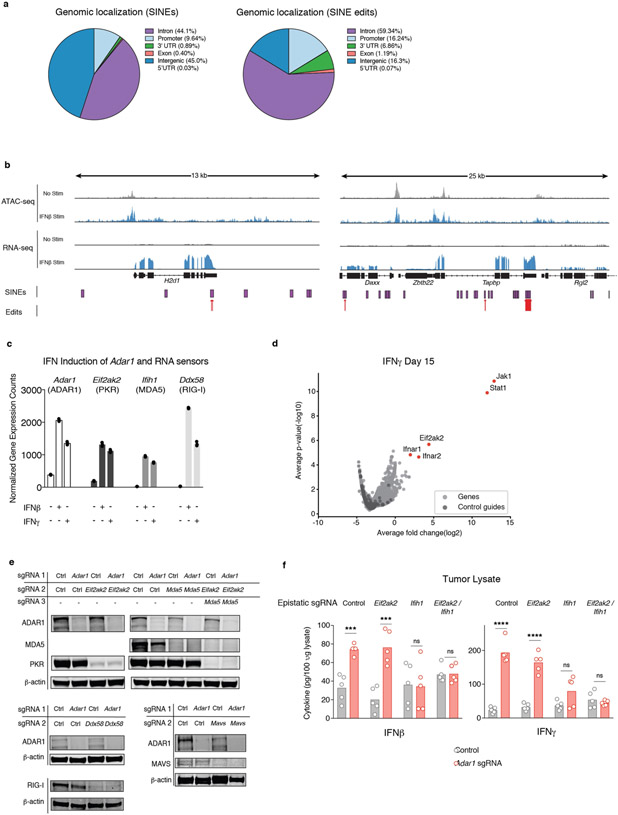
Extended Data Fig. 7∣. Corroborating data for dsRNA editing and epistasis studies of Adar1-null tumours.
a, Genomic localization of SINEs (left) and detected editing sites within SINEs (right) in control B16 tumour cells. b, Representative tracks of ATAC-seq and RNA-seq mapped to SINEs and detected edits in IFN-inducible regions of accessible chromatin and transcription. c, Transcriptional upregulation of Adar1 and dsRNA sensors 36 h after stimulation with IFNβ or IFNγ in control B16 tumour cells as measured by RNA-seq (n = 3 for each condition). d, Volcano plot depicting the relative depletion and enrichment of sgRNAs targeting 20,146 genes in a Cas9+ Adar1-null B16 tumour cell line following stimulation with IFNγ in vitro. P values are derived using STARS v1.3. e, Western blots demonstrating the loss of expression of PKR, MDA5, RIG-I, MAVS and ADAR1 from double knockout and triple knockout B16 tumour cell lines. Data are representative of two independent experiments. f, IFNβ and IFNγ ELISAs from tumour lysate extracted from Adar1-null and control tumours that were epistatically deleted for dsRNA sensors including Eif2ak2 (PKR), Ifih1 (MDA5) or both (n = 5 for each condition). f, Two-sided Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.001; ****p < 0.0001.