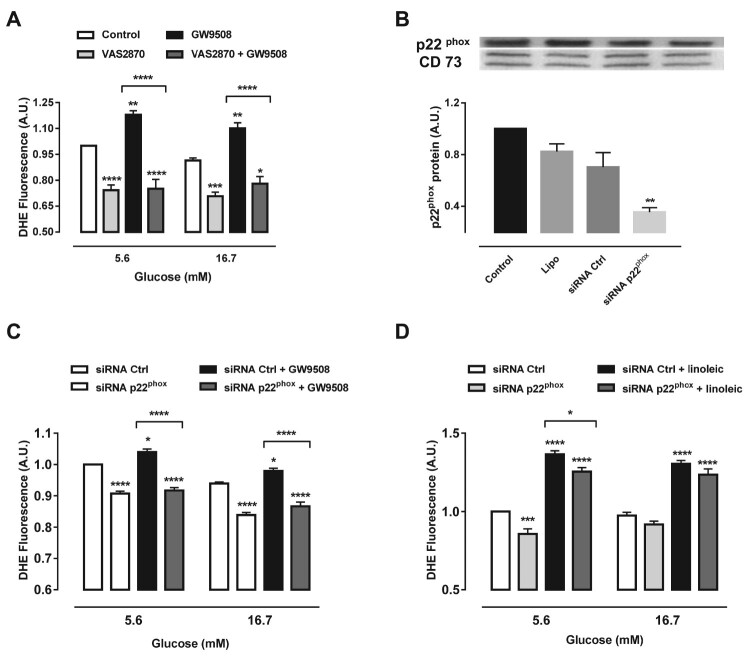
Figure 3.
Inhibition of NADPH oxidase reduces superoxide production induced by GW9508 and linoleic acid. (A) Production of superoxide in BRIN-BD11 cells after stimulation of GPR40 with 20 μM GW9508 in the presence or absence of a NADPH oxidase inhibitor, VAS2870, at different glucose concentrations (5.6 and 16.7 mM) for 60 min. Results are expressed as mean ± SEM of 7 independent experiments. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 versus Control or versus GW9508 (in brackets) in corresponding glucose concentration. Two-way ANOVA followed by Tukey. (B) Protein expression of p22phox in BRIN-BD11 cells submitted to small interfering RNA (siRNA) protocol. Conditions: non-transfected cells (Control), cells with lipofectamine alone (Lipo), cells transfected with scrambled siRNA (siRNA Ctrl) and cells transfected with siRNA p22phox (siRNA p22phox). Western blot analysis was performed using anti-p22phox and anti-CD73 (internal control). Results are expressed as mean ± SEM of 4 independent experiments. **P<0.01 versus siRNA Ctrl. One-way ANOVA followed by Sidak. (C) Production of superoxide in BRIN-BD11 cells transfected with scrambled siRNA (siRNA Ctrl) or siRNA p22phox in absence or presence of 20 μM GW9508 in 5.6 or 16.7 mM glucose for 60 min. Results are expressed as mean ± SEM of 13 independent experiments. *P<0.05 and ****P<0.0001 versus siRNA Ctrl or siRNA Ctrl + GW9508 (in brackets) in corresponding glucose concentration. Two-way ANOVA followed by Tukey. (D) Production of superoxide in BRIN-BD11 cells transfected with scrambled siRNA (siRNA Ctrl) or siRNA p22phox in absence or presence of 100 μM linoleic acid (Linoleic) in 5.6 or 16.7 mM glucose for 60 min. Results are expressed as mean ± SEM of 4 independent experiments. *P<0.05, ***P<0.001 and ****P<0.0001 versus siRNA Ctrl or siRNA Ctrl + linoleic (in brackets) in corresponding glucose concentration. Two-way ANOVA followed by Tukey. (A,C,D) Production of superoxide was analyzed by flow cytometry using DHE dye.