Figure 2. Treg cells from male and female VAT are distinct in their transcriptional profile and chromatin accessibility.
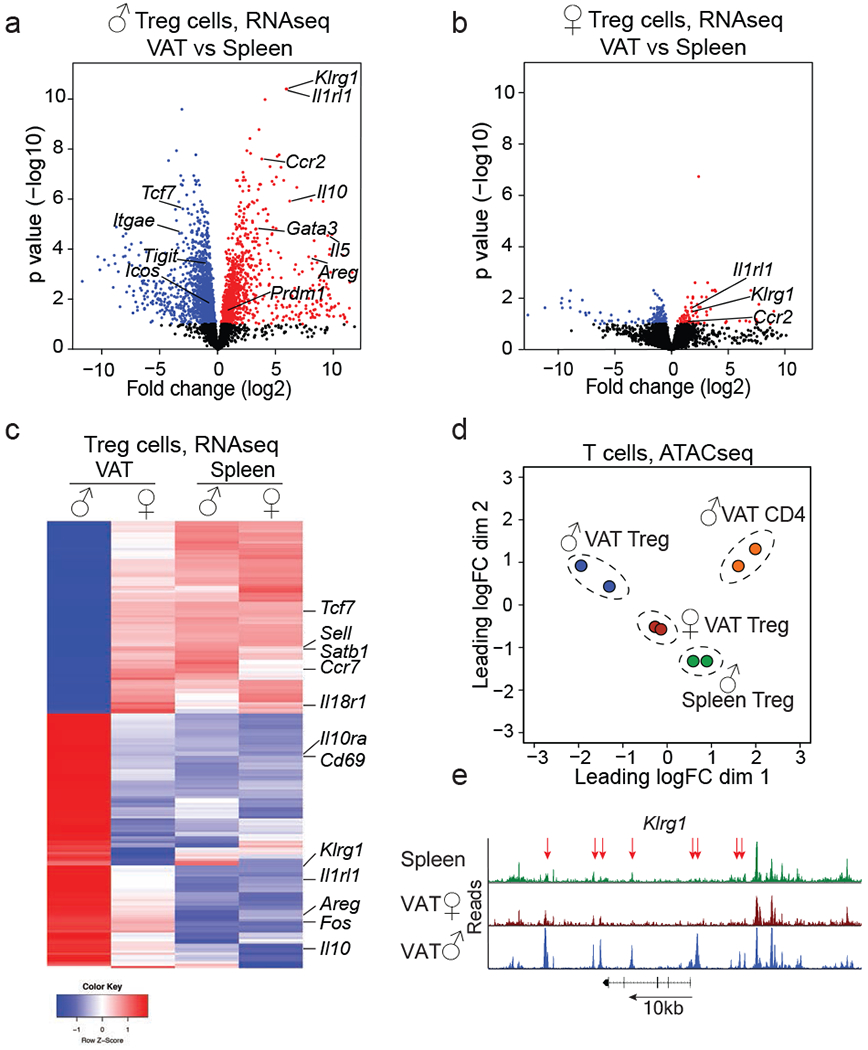
Treg cells were sorted from visceral adipose tissue (VAT) and spleens of 25 to 32-week-old Foxp3RFP mice to perform RNA sequencing and Assay for Transposase-Accessible Chromatin using sequencing (ATACseq). n=2 samples, each sample contains Treg cells from n=5 male and n=12 female mice. a, Volcano plot shows genes differentially expressed between Treg cells from male VAT and spleen. Each dot represents a gene; genes in red are up, genes in blue are down-regulated in male VAT Treg cells. b, Volcano plot shows genes differentially expressed between Treg cells from female VAT and spleen. Genes in red and blue are up or down-regulated, respectively, in female VAT Treg cells. c, Heatmap shows top 200 genes differentially expressed between male and female VAT and comparison to splenic Treg cells. d, Multi-dimensional scaling analysis of ATACseq data. Distances shown on the plot represent the leading log2-fold-change between samples. (e) ATACseq tracks show chromatin accessibility at the Klrg1 locus of male splenic Treg cells (green) and Treg cells from female (red) and male (blue) VAT. Arrows indicate regions of differential chromatin accessibility. Statistical methods and software packages described in methods.
