Abstract
During our investigation of Camellia sinensis diseases (2013–2018), a new leaf spot disease was found in seven provinces of China (Anhui, Fujian, Guangxi, Guizhou, Jiangxi, Tibet and Yunnan), occurring on both arboreal and terraced tea plants. The leaf spots were round to irregular, brown to dark brown, with grey or tangerine margins. Multi-locus (LSU, ITS, gapdh, tef-1α, tub2) phylogenetic analyses combined with morphological observations revealed four new species belonging to the genus Setophoma, i.e. S. antiqua, S. longinqua, S. yingyisheniae and S. yunnanensis. Of these four species, S. yingyisheniae was found to be present on diseased terraced tea plants in six of the seven sampled provinces (excluding Yunnan). The other three species only occurred on arboreal tea plants in Yunnan Province. In addition to the four species isolated from diseased leaves, S. endophytica sp. nov. was isolated from healthy leaves of terraced tea plants.
Keywords: five new taxa, fungal pathogen, phylogeny, taxonomy, tea plants
INTRODUCTION
During our investigation of diseases of tea plants (Camellia sinensis) cultivated in China in 2013, a new leaf spot disease was found to cause severe yield losses in Guangxi Province. The associated leaf spots were circular to irregular, brown to dark brown in colour, with grey or tangerine margins. A subsequent collection effort of similarly affected tea plant leaves has been ongoing in many tea plantations located in six provinces of China, i.e. Anhui, Fujian, Guizhou, Jiangxi, Tibet and Yunnan. Preliminary morphological observations and molecular analyses identified the associated fungi as Setophoma spp. To our knowledge, this is the first report of Setophoma on tea plants. Considering the potential commercial yield losses in tea plantations and the limited knowledge of this disease, accurate identification of the causal organisms is of great importance.
The genus Setophoma (Phaeosphaeriaceae) was introduced to accommodate Phoma terrestris and Pyrenochaeta sacchari (de Gruyter et al. 2010). Species of Setophoma are characterised as having setose pycnidia, phialidic conidiogenous cells and hyaline, ellipsoidal to subcylindrical, aseptate conidia (de Gruyter et al. 2010, Quaedvlieg et al. 2013). According to Index Fungorum and MycoBank, four additional Setophoma species have been described since the genus was introduced in 2010. The currently recognised species are: S. chromolaenae, S. cyperi, S. poaceicola, S. sacchari, S. terrestris, and S. vernoniae. All except S. terrestris are reported to occur on unique host plants (Table 1).
Table 1.
Host and distribution of Setophoma species.
| Species | Host | Distribution | References |
|---|---|---|---|
| S. antiqua | Camellia sinensis | China | This study |
| S. chromolaenae | Chromolaena odorata | Brazil | Quaedvlieg et al. (2013) |
| S. cyperi | Cyperus sphaerocephalus | South Africa, Eastern Cape | Crous et al. (2016) |
| S. endophytica | Camellia sinensis | China | This study |
| S. longinqua | Camellia sinensis | China | This study |
| S. poaceicola | Grass | Thailand | Thambugala et al. (2017) |
| S. sacchari | Saccharum officinarum | Brazil | de Gruyter et al. (2010) |
| S. terrestris | Allium cepa | North America, Senegal | de Gruyter et al. (2010) |
| Allium sativum | United States | de Gruyter et al. (2010) | |
| Brassica sp. | Canada, Alberta | Yang et al. (2017) | |
| Cucurbita maxima | USA, Oregon | Rivedal et al. (2018) | |
| Cucurbita moschata | Japan | Ikeda et al. (2012) | |
| Solanum lycopersicum | Canada, Ontario | Johnston-Monje et al. (2017) | |
| S. vernoniae | Vernonia polyanthes | Brazil | Crous et al. (2014) |
| S. yingyisheniae | Camellia sinensis | China | This study |
| S. yunnanensis | Camellia sinensis | China | This study |
The aim of the present study was to investigate the taxonomic and phylogenetic relationships of Setophoma spp. associated with tea plants based on multi-locus phylogenetic analyses, morphological comparison, host association and geographical distribution.
MATERIALS AND METHODS
Isolates
Isolates were obtained from either diseased or healthy tea plant tissues collected from 18 locations in seven provinces of China, following the single spore isolation and tissue isolation methods described in Liu et al. (2015). Type specimens of new species were deposited in the Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), with the ex-type living cultures being deposited in the China General Microbiological Culture Collection Center (CGMCC).
DNA extraction, PCR amplification and phylogenetic analyses
Total genomic DNA was extracted from fresh mycelia using the CTAB method. Five partial loci including the large subunit of the nrDNA (LSU), the 5.8S nuclear ribosomal RNA gene with the two flanking internally transcribed spacer regions (ITS), partial glyceraldehyde-3-phosphate dehydrogenase (gapdh), translation elongation factor 1-alpha (tef-1α) and β-tubulin (tub2) were amplified and sequenced using the following primer pairs: ITS1/ITS4 for ITS (White et al. 1990), LR0R/LR5 for LSU (Vilgalys & Hester 1990, Rehner & Samuels 1994), gpd1/gpd2 for gapdh (Berbee et al. 1999), EF-1/EF-2 for tef-1α (O’Donnell et al. 1998) and T1/Bt2b for tub2 (Glass & Donaldson 1995, O’Donnell & Cigelnik 1997). Amplicons for LSU, ITS, tef-1α and tub2 were generated according to Liu et al. (2019) while amplicons for gapdh were generated according to Liu et al. (2016). Amplicons were sequenced with both forward and reverse primers by the Omegagenetics Company, Beijing, China. MEGA v. 7.0.21 was used to generate consensus sequences.
DNA sequences (ITS and LSU) used to infer generic affiliation within the Phaeosphaeriaceae were downloaded from the NCBI database (Table 2). Sequence datasets of ITS, gapdh, tub2 and tef-1α (Table 3) were used for species delimitation in Setophoma. Sequence alignments were made using MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html) and were then manually edited in MEGA v. 7.0.21. Subsequently, individual alignments of above-mentioned loci were concatenated using Mesquite v. 3.4. The Maximum Likelihood (ML) and Bayesian analysis (BA) methods were used for phylogenetic inferences of both the single gene and concatenated alignments as described in Liu et al. (2019). The resulting trees were plotted using FIGTREE v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). All the alignments derived from this study were deposited in TreeBASE (https://treebase.org/) (S24160).
Table 2.
GenBank accession numbers from NCBI database.
| Name | Strain/specimen No.a | LSU | ITS |
|---|---|---|---|
| Acericola italica | MFLUCC 13-0609* | MF167429 | MF167428 |
| Allophaeosphaeria muriformia | MFLUCC 13-0349* | KP765681 | KP765680 |
| Allophaeosphaeria subcylindrospora | MFLUCC 13-0380* | KT314183 | KT314184 |
| Amarenomyces ammophilae | CBS 114595 | GU301859 | KF766146 |
| Ampelomyces quisqualis | CBS 129.79* | EU754128 | HQ108038 |
| Bhatiellae rosae | MFLUCC 17-0664* | MG828989 | MG828873 |
| Chaetosphaeronema achilleae | MFLUCC 16-0476 | KX765266 | KX765265 |
| Chaetosphaeronema hispidulum | CBS 216.75 | KF251652 | KF251148 |
| Coniothyrium carteri | CBS 105.91 | KF251712 | KF251209 |
| Dactylidina dactylidis | MFLUCC 14-0966* | MG829002 | MG828886 |
| Dematiopleospora rosicola | MFLU 16-0232* | MG829006 | MG828888 |
| Dematiopleospora salsolae | MFLUCC 17-0828* | MG829007 | MG828889 |
| Didymocyrtis consimilis | Gardiennet 12041 | KT383796 | KT383813 |
| Didymocyrtis ramalinae | Ertz 16399 | KT383802 | KT383838 |
| Embarria clematidis | MFLUCC 14-0976* | MG828987 | MG828871 |
| Galiicola pseudophaeosphaeria | MFLUCC 14-0524* | KT326693 | KT326692 |
| Hawksworthiana alliariae | MFLUCC 13-0070 | KX494877 | KX494876 |
| Hawksworthiana clematidicola | MFLUCC 14-0910* | MG829011 | MG828901 |
| Italica achilleae | MFLUCC 14-0959* | MG829013 | MG828903 |
| Juncaceicola achilleae | MFLUCC 13-0606* | KX449526 | KX449525 |
| Juncaceicola luzulae | MFLUCC 16-0780* | KX449530 | KX449529 |
| Juncaceicola typharum | CBS 296.54 | KF251695 | KF251192 |
| Leptospora galii | KUMCC 15-0521 | KX599548 | KX599547 |
| Leptospora rubella | CPC 11006 | DQ195792 | DQ195780 |
| Muriphaeosphaeria galatellae | MFLUCC 14-0614* | KT438329 | KT438333 |
| Neosetophoma samarorum | CBS 138.96* | KF251664 | KF251160 |
| Neostagonospora caricis | CBS 135092* | KF251667 | KF251163 |
| Neostagonospora elegiae | CBS 135101* | KF251668 | KF251164 |
| Neosulcatispora agaves | CPC 26407* | KT950867 | KT950853 |
| Nodulosphaeria hirta | MFLUCC 13-0867 | KU708845 | KU708849 |
| Nodulosphaeria scabiosae | MFLUCC 14-1111* | KU708846 | KU708850 |
| Ophiobolopsis italica | MFLUCC 17-1791* | MG520959 | MG520939 |
| Ophiobolus artemisiae | MFLUCC 14-1156* | KT315509 | KT315508 |
| Ophiobolus disseminans | MFLUCC 17-1787* | MG520961 | MG520941 |
| Ophiosimulans tanaceti | MFLUCC 14-0525 | KU738891 | KU738890 |
| Paraophiobolus arundinis | MFLUCC 17-1789* | MG520965 | MG520945 |
| Paraophiobolus plantaginis | MFLUCC 17-0245* | KY815010 | KY797641 |
| Paraphoma chrysanthemicola | CBS 172.70 | KF251669 | KF251165 |
| Paraphoma radicina | CBS 102875 | KF251677 | KF251173 |
| Paraphoma rhaphiolepidis | CBS 142524* | KY979813 | KY979758 |
| Paraphoma vinacea | UMPV001 = BRIP 63684 | KU176888 | KU176884 |
| Parastagonospora caricis | CBS 135671 | KF251680 | KF251176 |
| Parastagonospora nodorum | CBS 110109 | KF251681 | KF251177 |
| Parastagonospora poagena | CBS 136776* | KJ869174 | KJ869116 |
| Parastagonosporella fallopiae | CBS 135981* | MH460545 | MH460543 |
| CCTU 1151.1 | MH460546 | MH460544 | |
| Phaeosphaeria oryzae | CBS 110110* | KF251689 | KF251186 |
| Phaeosphaeria papayae | CBS 135416 | KF251690 | KF251187 |
| Phaeosphaeriopsis glaucopunctata | CBS 653.86 | KF251702 | KF251199 |
| Poaceicola arundinis | MFLU 15-0702* | KU058726 | KU058716 |
| Poaceicola italica | MFLUCC 13-0267* | KX910094 | KX926421 |
| Populocrescentia forlicesenensis | MFLU 15-0651* | KT306952 | KT306948 |
| Pseudoophiobolus achilleae | MFLU 17-0925* | MG520966 | MG520946 |
| Pseudoophiobolus mathieui | MFLUCC 17-1784 | MG520969 | MG520949 |
| Pseudophaeosphaeria rubi | MFLUCC 14-0259* | KX765299 | KX765298 |
| Sclerostagonospora rosicola | MFLUCC 15-0129* | MG829068 | MG828957 |
| Septoriella allojunci | MFLU 15-0701* | KU058728 | KU058718 |
| Septoriella phragmitis | CPC 24118 = CBS 140065* | KR873279 | KR873251 |
| Setophoma chromolaenae | CBS 135105* | KF251747 | KF251244 |
| Setophoma cyperi | CPC 25702 = CBS 141450* | KX228337 | KX228286 |
| Setophoma poaceicola | MFLUCC 16-0880* | KY550386 | KY568988 |
| Setophoma sacchari | MFLUCC 12-0241 | KJ476147 | KJ476145 |
| CBS 333.39* | MH867535 | MH856038 | |
| Setophoma terrestris | CBS 335.87 | KF251750 | KF251247 |
| CBS 377.52 | KF251751 | KF251248 | |
| Setophoma vernoniae | CPC 23123 = CBS 137988* | KJ869198 | KJ869141 |
| Sulcispora pleurospora | MFLUCC 14-0995* | KP271444 | KP271443 |
| Tintelnotia destructans | CBS 127737* | KY090664 | KY090652 |
| Tintelnotia opuntiae | CBS 376.91* | GU238123 | KY090651 |
| Vagicola vagans | CBS 604.86 | KF251696 | KF251193 |
| Wojnowicia lonicerae | MFLUCC 13-0737* | KP684151 | KP744471 |
| Wojnowicia rosicola | MFLUCC 15-0128* | MG829091 | MG828979 |
| Wojnowiciella eucalypti | CPC 25024* | KR476774 | KR476741 |
| Xenoseptoria neosaccardoi | CBS 120.43 | KF251783 | KF251280 |
| CBS 128665* | KF251784 | KF251281 |
aEx-type strains are marked with asterisk *.
Table 3.
Collection details and GenBank accession numbers of isolates included in this study.
| Species | Strain No.a | Habitat | Location |
GenBank accession no.b |
||||
|---|---|---|---|---|---|---|---|---|
| LSU | ITS | tub2 | tef-1α | GAPDH | ||||
| Didymella pinodella | CBS 531.66 | Trifolium pretense | USA | GU238017 | FJ427052 | FJ427162 | MK525067 | MK532379 |
| Setophoma antiqua | LC6594 | Camellia sinensis, pathogen | Yunnan, China | MK511947 | MK511909 | MK524999 | MK525070 | MK525034 |
| LC6595 | Camellia sinensis, pathogen | Yunnan, China | MK511948 | MK511910 | MK525000 | MK525071 | MK525035 | |
| CGMCC 3.19525 = LC6596* | Camellia sinensis, pathogen | Yunnan, China | - | MK511911 | MK525001 | MK525072 | MK525036 | |
| Setophoma chromolaenae | CBS 135105* | Chromolaena odorata | Brazil | KF251747 | KF251244 | KF252728 | KF253195 | - |
| Setophoma endophytica | LC13538 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511923 | MK525012 | MK525084 | MK525047 |
| LF2067 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511924 | MK525013 | MK525085 | MK525048 | |
| CGMCC 3.19528 = LC3163* | Camellia sinensis, endophyte | Jiangxi, China | MK511956 | MK511931 | MK525020 | MK525092 | MK525053 | |
| LC3164 | Camellia sinensis, endophyte | Jiangxi, China | MK511957 | MK511932 | MK525021 | MK525093 | MK525054 | |
| LC3165 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511933 | MK525022 | MK525094 | MK525055 | |
| LC3216 | Camellia sinensis, endophyte | Jiangxi, China | MK511959 | MK511938 | MK525026 | MK525099 | MK525060 | |
| LC3297 | Camellia sinensis, endophyte | Jiangxi, China | MK511962 | MK511941 | MK525029 | MK525102 | MK525063 | |
| Setophoma longinqua | CGMCC 3.19524 = LC6593* | Camellia sinensis, pathogen | Yunnan, China | MK511946 | MK511908 | MK524998 | MK525069 | - |
| LC13481 | Camellia sinensis, pathogen | Yunnan, China | - | MK511925 | MK525014 | MK525086 | - | |
| LC13482 | Camellia sinensis, pathogen | Yunnan, China | - | MK511926 | MK525015 | MK525087 | - | |
| Setophoma sp. | LC12841 | Carbonatite in cave | Guizhou, China | - | MK511927 | MK525016 | MK525088 | MK525049 |
| LC12842 | Carbonatite in cave | Guizhou, China | - | MK511928 | MK525017 | MK525089 | MK525050 | |
| CGMCC 3.19526 = LC7511 | Carbonatite in cave | Guizhou, China | MK511965 | MK511944 | MK525032 | MK525105 | MK525066 | |
| Setophoma terrestris | CBS 335.29 = MUCL 9892 = LC6449* | Allium sativum | USA | KF251749 | KF251246 | KF252729 | KF253196 | - |
| CBS 335.87 | Allium cepa | Senegal | KF251750 | KF251247 | KF252730 | KF253197 | - | |
| CBS 377.52 | Allium cepa | - | KF251751 | KF251248 | KF252731 | KF253198 | - | |
| Setophoma yingyisheniae | LC6739 | Camellia sinensis, pathogen | Guizhou, China | - | MK511912 | MK525002 | MK525073 | - |
| LC12696 | Camellia sinensis, pathogen | Anhui, China | MK511950 | MK511914 | - | MK525075 | MK525038 | |
| LC12699 | Camellia sinensis, pathogen | Anhui, China | MK511951 | MK511915 | MK525004 | MK525076 | MK525039 | |
| LC13477 | Camellia sinensis, pathogen | Fujian, China | MK511952 | MK511916 | MK525005 | MK525077 | MK525040 | |
| LC13478 | Camellia sinensis, pathogen | Fujian, China | - | MK511917 | MK525006 | MK525078 | MK525041 | |
| CGMCC 3.19527 = LC13479* | Camellia sinensis, pathogen | Guangxi, China | - | MK511918 | MK525007 | MK525079 | MK525042 | |
| LC13480 | Camellia sinensis, pathogen | Guangxi, China | MK511953 | MK511919 | MK525008 | MK525080 | MK525043 | |
| LF2026 | Camellia sinensis, pathogen | Guangxi, China | - | MK511920 | MK525009 | MK525081 | MK525044 | |
| LF2027 | Camellia sinensis, pathogen | Guangxi, China | MK511954 | MK511921 | MK525010 | MK525082 | MK525045 | |
| LF2032 | Camellia sinensis, pathogen | Guangxi, China | - | MK511922 | MK525011 | MK525083 | MK525046 | |
| LC3133 | Camellia sinensis, pathogen | Jiangxi, China | MK511955 | MK511929 | MK525018 | MK525090 | MK525051 | |
| LC3137 | Camellia sinensis, pathogen | Jiangxi, China | - | MK511930 | MK525019 | MK525091 | MK525052 | |
| LC3176 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511934 | - | MK525095 | MK525056 | |
| LC3181 | Camellia sinensis, endophyte | Jiangxi, China | MK511958 | MK511935 | MK525023 | MK525096 | MK525057 | |
| LC3185 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511936 | MK525024 | MK525097 | MK525058 | |
| LC3197 | Camellia sinensis, endophyte | Jiangxi, China | - | MK511937 | MK525025 | MK525098 | MK525059 | |
| LC3236 | Camellia sinensis, pathogen | Jiangxi, China | MK511960 | MK511939 | MK525027 | MK525100 | MK525061 | |
| LC3276 | Camellia sinensis, pathogen | Jiangxi, China | MK511961 | MK511940 | MK525028 | MK525101 | MK525062 | |
| LC3334 | Camellia sinensis, pathogen | Jiangxi, China | MK511963 | MK511942 | MK525030 | MK525103 | MK525064 | |
| LC3499 | Camellia sinensis, endophyte | Guangxi, China | MK511964 | MK511943 | MK525031 | MK525104 | MK525065 | |
| Setophoma yunnanensis | LC6532 | Camellia sinensis, pathogen | Yunnan, China | MK511945 | MK511907 | MK524997 | MK525068 | MK525033 |
| CGMCC 3.19529 = LC6753* | Camellia sinensis, pathogen | Yunnan, China | MK511949 | MK511913 | MK525003 | MK525074 | MK525037 | |
aEx-type strains are marked with star *; CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection Center, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; LC: working collection of Lei Cai, housed at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; MUCL: Mycothèque de l’Université catholique de Louvain, Louvain-la-Neuve, Belgium.
bNewly generated sequences are indicated in bold.
Morphology
Cultures were cultivated on both 2 % malt extract agar (MEA) and potato dextrose agar (PDA) (DifcoTM, Becton, Dickinson and Company, Sparks, MD, USA) at 25 °C in a 12 h day/night regime. Growth rates were measured after 10 d in the dark. Colony colours were rated following the colour charts of Rayner (1970). In order to observe wall structures of mature conidiomata, sections were made using a Leica CM1950 freezing microtome. Morphological observations of reproductive structures were made in lactic acid and observed using a Nikon Eclipse 80i microscope using differential interference contrast (DIC) illumination. At least 30 measurements per structure were taken and the mean value, standard deviation and minimum–maximum values are given, with the extreme measurements given in parentheses.
RESULTS
DNA phylogeny
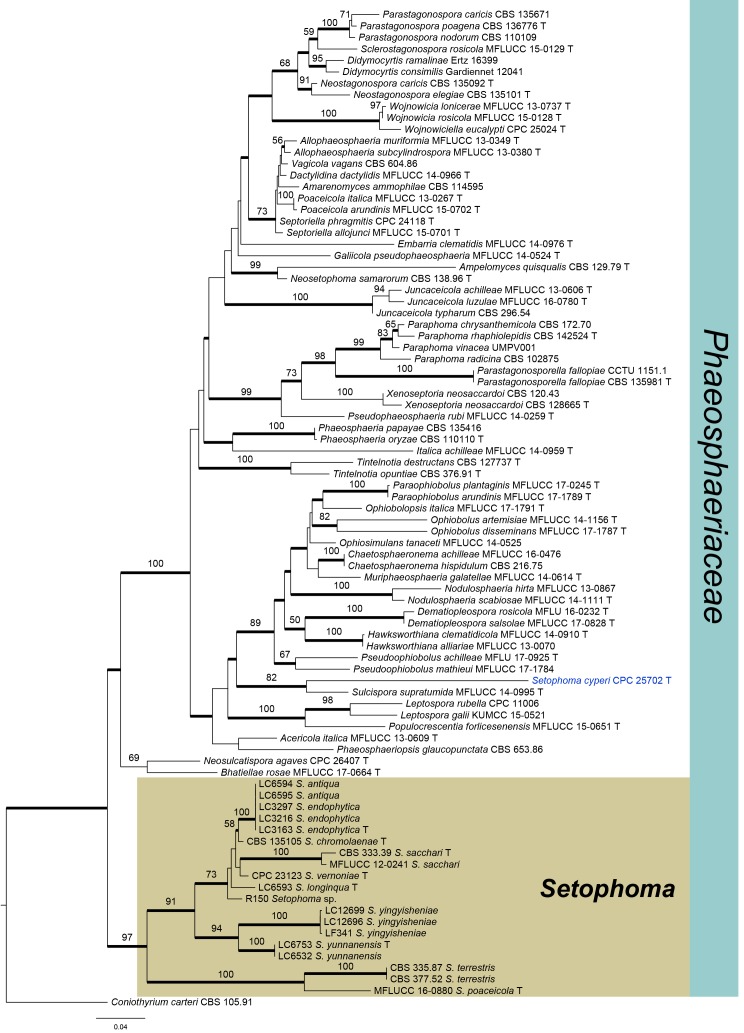
The concatenated alignment of LSU and ITS sequences comprised a total length of 1 397 characters (LSU: 818, ITS: 579) including alignment gaps, 486 of which were unique site patterns. The ML search resolved a best tree with a InL of -14098.620225. Fixed base frequency for LSU and ITS was set in the Bayesian analysis (BA). The BA lasted for 1 305 000 generations and the 50 % consensus tree and posterior probabilities were calculated from 3 908 trees, generated during two runs. The ML tree confirmed the same tree topology as that generated by the Bayesian analysis and it also revealed that strains isolated in this study clustered together with the generic type of Setophoma (S. terrestris), in the basal clade of Fig. 1. However, the ex-type strain of another known Setophoma species, S. cyperi (CPC 25702 = CBS 141450) clustered outside Setophoma s. str., and was more closely related to Sulcispora pleurospora (MFLUCC 14-0995) (Fig. 1).
Fig. 1.
Overview phylogenetic tree of Phaeosphaeriaceae (50 % majority rule consensus) resulting from a Bayesian analysis of the combined LSU and ITS sequence alignment. Bayesian posterior probabilities (PP > 0.95) are emphasized by thickened branches, maximum likelihood bootstrap support values (≥ 50 %) are shown at the nodes. The scale bar represents the expected number of changes per site. Ex-type cultures are indicated with “T” behind the taxa labels. The tree was rooted to Coniothyrium carteri (CBS 105.91).
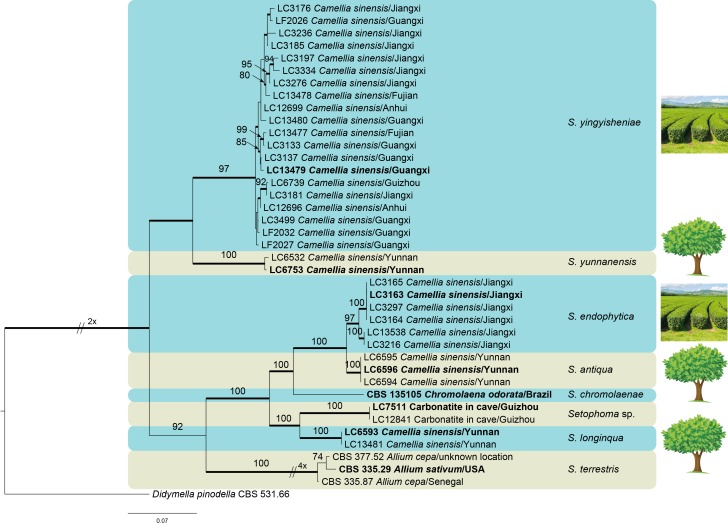
To better infer species delimitation in Setophoma, sequences of four loci (ITS, tef-1α, gapdh and tub2) were concatenated for further phylogenetic analyses. Strains of S. sacchari, S. vernoniae and S. poaceicola were excluded from this analysis due to their incomplete sequence datasets. The final alignment contained a total of 2 504 characters (ITS: 524, tef-1α: 875, gapdh: 550, tub2: 539) including alignment gaps, of which 872 were unique site patterns. The ML search revealed a best tree with an InL of -10791.633622. Fixed base frequency was set in the Bayesian analysis. The BA lasted for 1 315 000 generations and the 50 % consensus tree and posterior probabilities were calculated from 1 656 trees generated during two runs. The topology of the phylogenetic trees generated from both ML and BA methods were congruent (Fig. 2). Strains associated with tea plants clustered in five different clades.
Fig. 2.
Phylogenetic tree of Setophoma resulting from a Bayesian analysis of the combined ITS, gapdh, tef-1α and tub2 sequence alignment. Bayesian posterior probabilities (PP > 0.95) are emphasized by thickened branches, maximum likelihood bootstrap support values (> 50 %) are shown at the nodes. The scale bar represents the expected number of changes per site. The taxa names consist of strain number, host or substrate, and location. Ex-type strains are represented in bold. Setophoma yingyisheniae and S. endophytica were isolated from terraced tea plants, S. yunnanensis, S. antiqua and S. longinqua were isolated from arboreal tea plants (illustrated by photos on the right side of the figure). The tree was rooted to Didymella pinodella (CBS 531.66).
Taxonomy
A total of 94 isolates from diseased leaves and 17 isolates from healthy leaves were isolated from arboreal and terraced tea plants. Based on ITS sequence similarity and colonial morphology, 35 representative isolates were selected for further microscopic observation and phylogenetic analyses. These 35 strains formed five distinct and novel phylogenetic clades representing five new species (Fig. 2). These are described and illustrated below.
Setophoma Gruyter, Aveskamp & Verkley, Mycologia 102: 1077. 2010. emend. F. Liu & L. Cai.
Ascomata immersed or semi-immersed, uniloculate, globose to subglobose, brown to dark brown, solitary or gregarious, centrally ostiolate, papillate, wall of textura angularis or prismatica. Pseudoparaphyses hyaline, frequently septate. Asci bitunicate, fissitunicate, cylindrical to cylindrical-clavate, smooth-walled. Ascospores fusiform with rounded ends, hyaline, overlapping or irregularly biseriate, septate, usually widest at the second cell from apex, smooth-walled, guttulate. Conidiomata pycnidial, solitary to confluent, on upper surface or submerged in agar, globose to subglobose, setose or not, with papillate ostioles, honey to olivaceous or olivaceous black; pycnidial wall of pseudoparenchymatal cells. Conidiophores reduced to conidiogenous cells lining inner cavity, or simply branched. Conidiogenous cells hyaline, smooth, phialidic, discrete. Conidia aseptate, globose, subglobose, ellipsoidal to subcylindrical to subfusoid, guttulate.
Type species: S. terrestris (H.N. Hansen) Gruyter et al.
Notes: The genus Setophoma was introduced to accommodate phoma-like species with setose conidiomata by de Gruyter et al. (2010). However, the new species of S. antiqua, S. endophytica and S. yunnanensis, proposed in this study, lack setae. We therefore broadened the generic concept of the genus Setophoma to also accommodate species lacking setose conidiomata.
Setophoma antiqua F. Liu & L. Cai, sp. nov. MycoBank MB829900. Fig. 3.
Fig. 3.
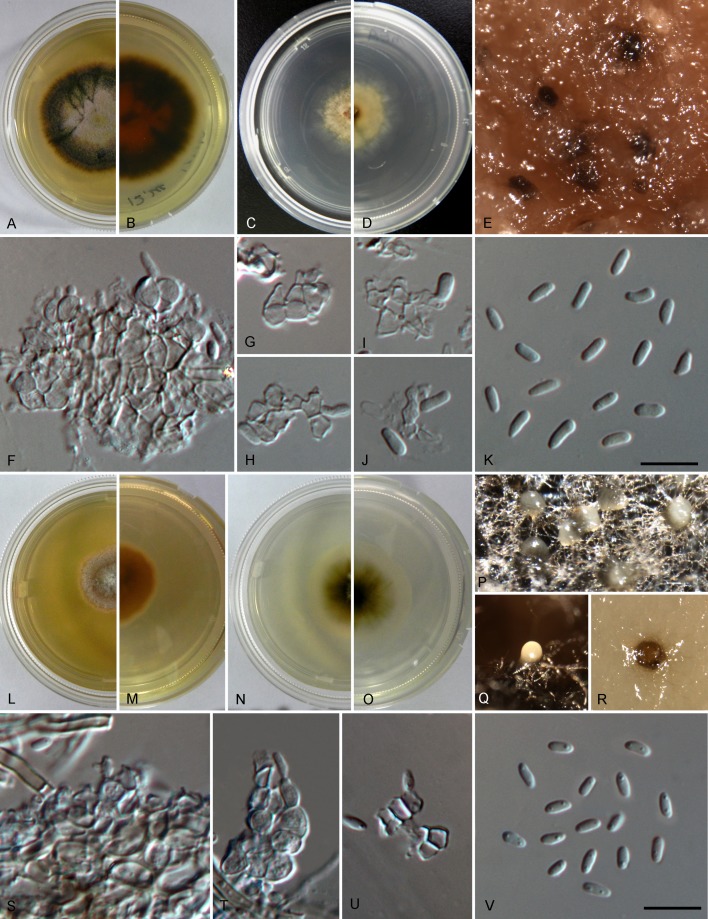
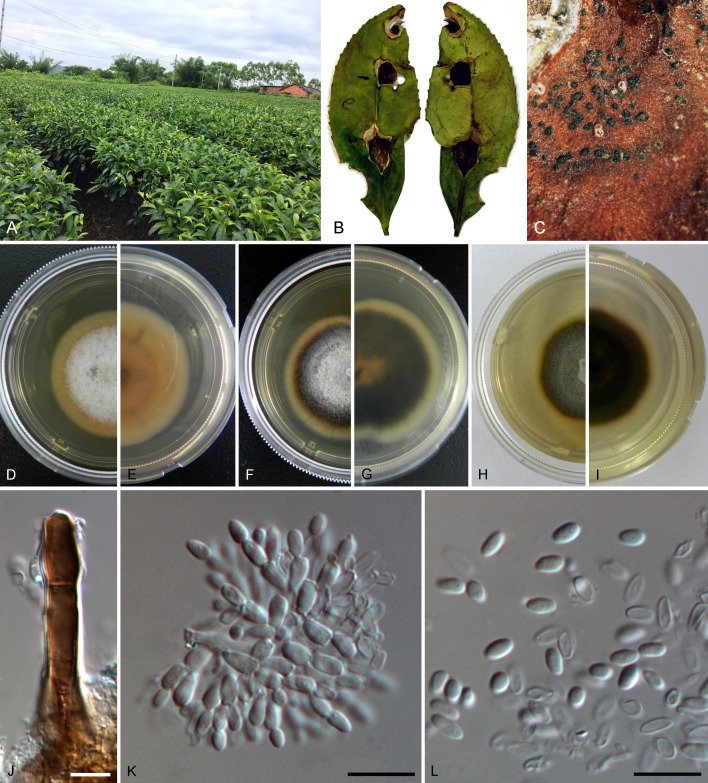
Setophoma antiqua (ex-type CGMCC 3.19525 = LC6596). A. Sampling environment. B. Symptom of diseased leaf. C, D. Front and back of colony on PDA. E, F. Front and back of colony on MEA. G. Conidiomata. H–J. Vertical sections of conidiomata. K. Conidiogenous cells. L, M. Conidia. Scale bars = 10 μm.
Etymology: From the Latin “antiquus” = old, referring to its host, old tea plant.
Sexual morph: Unknown. Asexual morph: Aerial mycelia hyaline or pale brown, branched, septate. Conidiomata pycnidial, scattered or gregarious, immersed or semi-immersed, globose to subglobose, olivaceous, 100–250 μm diam. Pycnidial wall pale to dark brown, 4–15 μm wide, with 3–4 layers, walls of textura angularis. Conidiophores reduced to conidiogenous cells lining inner cavity. Conidiogenous cells single, phialidic, unbranched, aseptate, hyaline, smooth, ampulliform, 3–5.5 × 2.5–5 μm (av. = 3.8 ± 0.6 × 3.2 ± 0.7 μm). Conidia hyaline, aseptate, ellipsoid with rounded ends or allantoid, smooth-walled, 2.5–5 × 1.5–2.5 μm (av. = 3.7 ± 0.5 × 1.8 ± 0.2 μm).
Culture characteristics: On PDA, umbonate with entire edge, luteous in the centre, grey to deep grey at the edge, reverse deep grey, reaching 33–36 mm diam after 10 d at 25 °C. On MEA, flat with entire edge, front and reverse pale olivaceous in the centre and pale grey at the edge, reaching 24–29 mm diam after 10 d at 25 °C.
Typus: China, Yunnan Province, Xishuangbanna, Mengla County, on old/arboreal Camellia sinensis, 18 Apr. 2015, F. Liu (holotype HMAS 248083, culture ex-type CGMCC 3.19525 = LC6596 = LF1239).
Additional materials examined: China, Yunnan Province, Xishuangbanna, Mengla County, on old/arboreal Camellia sinensis, 18 Apr. 2015, F. Liu, living cultures LC6594 = LF1237, LC6595 = LF1238.
Notes: Based on the multi-locus phylogeny (Fig. 2), S. antiqua is phylogenetically related to S. endophytica and has high LSU (100 %), ITS (99 %) and gapdh (99 %) sequence similarities with the later. However, both species can be well differentiated from each other based on their tub2 (502/518, 97 % sequence similarity) and tef-1α (561/613, 92 % sequence similarity) sequences. In addition, the habitat of both species are different. Setophoma antiqua was isolated from diseased leaf spots of arboreal tea plant from an unmanaged forest on the mountain in Yunnan Province, while S. endophytica was isolated from healthy leaves of terraced tea plant from an open national forestry park in Jiangxi Province.
Setophoma endophytica F. Liu & L. Cai, sp. nov. MycoBank MB829902. Fig. 4.
Fig. 4.
Setophoma endophytica (A–K. ex-type CGMCC 3.19528 = LC3163, L–V. LC3216). A, B, L, M. Front and back of colonies on PDA. C, D, N, O. Front and back of colonies on MEA. E, P–R. Conidiomata. F–J, S–U. Conidiogenous cells and conidia. K, V. Conidia. Scale bars = 10 μm.
Etymology: Named after its original habitat as an endophyte.
Sexual morph: Unknown. Asexual morph: Aerial mycelia hyaline, pale brown, smooth, branched, septate. Conidiomata immersed, olivaceous, globose to subglobose, scattered, 109–200 μm diam. Pycnidial wall pale brown, with 4–5 layers, 15–25 μm wide, walls of textura angularis. Conidiophores reduced to conidiogenous cells lining inner cavity. Conidiogenous cells hyaline, aseptate, smooth, ampulliform, rarely irregular, 3.5–6.5 × 3–5.5 μm (av. = 4.9 ± 0.4 × 3.6 ± 0.3 μm). Conidia hyaline, aseptate, smooth, cylindrical to reniform, with rounded ends, straight or slightly curved, 4–5.5 × 1.5–2 μm (av. = 4.8 ± 0.2 × 1.7 ± 0.1 μm).
Culture characteristics: On PDA, low convex with entire edge, pale grey in the centre and olivaceous at the edge, reverse dark brown in the centre and black at the edge, reaching 28–30 mm diam after 10 d at 25 °C. On MEA, umbonate with entire edge, front and reverse brown in the centre and buff at the edge, reaching 22–25 mm diam after 10 d at 25 °C.
Typus: China, Jiangxi Province, Ganzhou, Yangling National Forest Park, on healthy leaves of Camellia sinensis, 24 Apr. 2013, F. Liu, YLBE3 (holotype HMAS 248081, culture ex-type CGMCC 3.19528 = LC3163 = LF372).
Additional materials examined: China, Jiangxi Province, Ganzhou, Yangling National Forest Park, on healthy leaves of Camellia sinensis, 24 Apr. 2013, F. Liu, YLBE3, living cultures LC3164 = LF373, LC3265 = LF374, LC3297 = LF519; on healthy leaves of Camellia sinensis, 24 Apr. 2013, F. Liu, YLBE1, living cultures LC3216 = LF428, LC13538 = LF2066.
Notes: Setophoma endophytica was isolated from healthy leaves of terraced tea plants from an open national forestry park in Jiangxi Province. Whether it is actually pathogenic to tea plant needs further research. Although strains of S. endophytica form two subclades in the multi-locus phylogeny (Fig. 2), they were recognised as one species because of their same geographical origin, similar morphological characters, and high sequence similarity (100 % in ITS, 99 % in gapdh, 98 % in tef-1α and tub2). For the differences between S. endophytica and its closely related species, see the notes under S. antiqua.
Setophoma longinqua F. Liu & L. Cai, sp. nov. MycoBank MB829904. Fig. 5.
Fig. 5.
Setophoma longinqua (ex-type CGMCC 3.19524 = LC6593). A. Symptom of diseased leaf. B, C. Front and back of colony on PDA. D, E. Front and back of colony on MEA. F. Setae. G, H. Conidiogenous cells and conidia. I. Conidia. Scale bars = 10 μm.
Etymology: From the Latin longinqua = remote, refers to the remote mountain where the species was collected.
Sexual morph: Unknown. Asexual morph: Aerial mycelia hyaline or grey-brown, smooth, branched. Conidiomata pycnidial, subglobose to ovoid, with an opening at apex, 100–200 μm diam. Pycnidial wall pale brown, with 2–3 layers, walls of textura angularis. Setae erect, solitary, unbranched, aseptate or septate, smooth, medium-brown to brown, subcylindrical, apex rounded. Conidiophores reduced to conidiogenous cells lining the inner cavity. Conidiogenous cells phialidic, unbranched, aseptate, hyaline, ampulliform, smooth, 3.5–4.5 × 3–4 μm (av. = 3.9 ± 0.2 × 3.6 ± 0.2 μm). Conidia aseptate, smooth, hyaline, with small guttules, cylindrical or subcylindrical with round or obtuse ends, sometimes allantoid, 4–5.5 × 1.5–2 μm (av. = 4.9 ± 0.3 × 1.8 ± 0.2 μm).
Culture characteristics: On PDA, flat with undulate edge, luteous coloured in the centre and brown at the edge, reverse sepia, reaching 27–28 mm diam after 10 d at 25 °C. On MEA, flat with roughly entire edge, surface and reverse isabelline, reaching 21–22 mm diam after 10 d at 25 °C.
Typus: China, Yunnan Province, Xishuangbanna, Mengla County, on Camellia sinensis, 18 Apr. 2015, F. Liu, GFZCWS001P (holotype HMAS 248082, culture ex-type CGMCC 3.19524 = LC6593 = LF1236).
Additional materials examined: China, Yunnan Province, Xishuangbanna, Mengla County, on Camellia sinensis, 18 Apr. 2015, F. Liu, GFZCWS001P, living cultures LC13481 = LF2068, LC13482 = LF2069.
Notes: Setophoma longinqua was isolated from the same symptomatic leaf as S. antiqua, but they are phylogenetically distinct (Figs 1, 2). It is morphologically distinct from S. antiqua by producing seta and longer conidia (av. = 4.9 × 1.8 μm vs. 3.7 × 1.8 μm). Based on the multi-locus phylogeny (Fig. 2), the most closely related species to S. longinqua is an undescribed carbonatite associated species, but only with low sequence similarities of 95 % on ITS, 92 % on tef-1α, and 90 % on tub2.
Setophoma yingyisheniae F. Liu & L. Cai, sp. nov. MycoBank MB829903. Fig. 6.
Fig. 6.
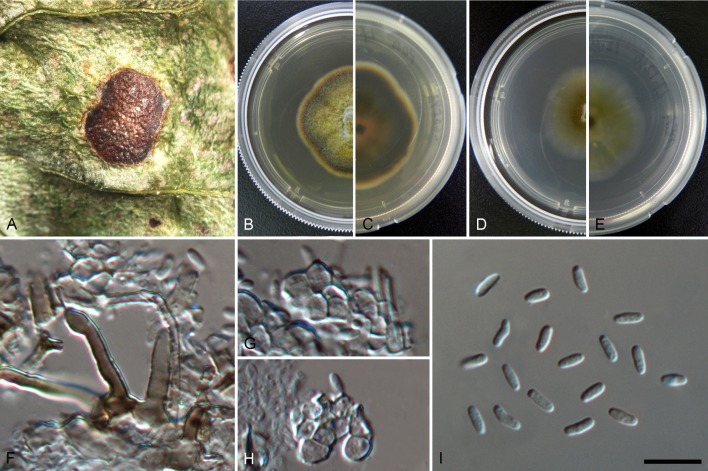
Setophoma yingyisheniae (B, C, J–L. holotype GXGL01a, D, E. LC3236, F, G. LC3334, H, I. LC3133). A. Representative sampling environment-terraced tea plant. B. Front and back of diseased leaves. C. Conidiomata on leaf. D–I. Front and back of colonies on PDA. J. Seta. K. Conidiophores, conidiogenous cells and conidia. L. Conidia. Scale bars = 10 μm.
Etymology: Named after the collector, Yingyi Shen, who was also the first to report Setophoma leaf spot disease on tea plants.
Sexual morph: Unknown. Sterile on cultural media. Lesions on plant leaves subglobose or irregular, initially off-white to brownish yellow, becoming dark grey in the centre, separated from the healthy tissue by a black margin. Conidiomata pycnidial, dark brown or black, globose or subglobose, with an opening at apex, 60–200 μm diam. Pycnidial wall brown, walls of textura angularis. Setae 8.5–10 μm wide, erect, solitary, unbranched, septate, dark brown, apex rounded. Conidiophores hyaline, often with two branches. Conidiogenous cells hyaline, oblong, 3–5.5 × 1.5–2.5 μm (av. = 4.2 ± 0.6 × 2.1 ± 0.2 μm). Conidia hyaline, aseptate, smooth, subglobose, reniform, or cylindrical, 3.5–6 × 1.5–2.5 μm (av. = 4.4 ± 0.5 × 2.2 ± 0.2 μm).
Typus: China, Guangxi Province, Guilin, Longsheng County, altitude 1 200 m, on Camellia sinensis, 21 Sep. 2016, Y.Y. Shen, GXGL01a (holotype HMAS 248086, culture ex-type CGMCC 3.19527 = LC13479 = LF1986).
Additional materials examined: China, Anhui Province, Bengbu County, Jianping Mountain, on Camellia sp., 2015, F. Liu, AHTEA07, living culture LC12696 = LF1529; on Camellia sp., 2015, F. Liu, AHTEA08, living culture LC12699 = LF1532; Fujian Province, Wuyishan City, Xingtianpu, 27.31′25″N, 118.1′59″E, 160 m, on C. sinensis, 22 Aug. 2016, F. Liu, WYSH3, living culture LC13477 = LF1935; on C. sinensis, 22 Aug. 2016, F. Liu, WYSH4, living culture LC13478 = LF1943; Guangxi Province, Guilin, Longsheng County, altitude 1 200 m, on Camellia sinensis, 21 Sep. 2016, Y.Y. Shen, GXGL02, living cultures LC13480 = LF2017, LF2026; on C. sinensis, 21 Sep. 2016, Y.Y. Shen, GXGL04, living cultures LF2027, LF2032; Guilin, Tea Science and Research Institute of GuiLin, on healthy twig of Camellia sp., Sep. 2013, T.W. Hou, living culture LC3499 = LF727; Guizhou Province, on Camellia sp., 2015, F. Liu, GZSXD001P, living culture LC6739 = LF1420; Jiangxi Province, Ganzhou City, Yangling National Forest Park, on C. sinensis, 24 Apr. 2013, F. Liu, YLA2, living culture LC3133 = LF341; on healthy leaves of C. sinensis, 24 Apr. 2013, F. Liu, YLBE5, living culture LC3176 = LF386; Nanchang City, Meiling, on healthy leaves of C. sinensis, Apr. 2013, F. Liu, MLE002, living culture LC3197 = LF407.
Notes: Setophoma yingyisheniae was isolated from both symptomatic and asymptomatic leaves of terraced (shrubby) tea plants from five provinces, i.e. Anhui, Fujian, Guangxi, Guizhou and Jiangxi. It is phylogenetically related to S. yunnanensis (Fig. 2, 96 % sequence similarity on ITS, 88 % on gapdh, 87 % on tef-1α and 93 % on tub2), but differs from the later in the host (terraced vs. arboreal tea plant) and habitat (plantation vs. unmanaged forest, Figs 6A, 7B). With respect to the conidiogenesis, the conidiophores of S. yunnanensis are simple and often reduced to single and ampulliform or globose conidiogenous cells, while these are branched in S. yingyisheniae and produces oblong conidiogenous cells. In addition, S. yingyisheniae differs from S. yunnanensis by producing seta in culture.
Fig. 7.
Setophoma yunnanensis (ex-type CGMCC 3.19529 = LC6753). A, B. Sampling environment and arboreal tea plants. C, D. Symptom of diseased leaves. E, F. Front and back of colony on PDA. G, H. Front and back of colony on MEA. I–K. Vertical section of conidiomata. L. Conidiogenous cell and conidium. M, N. Conidia. Scale bars = 10 μm.
Setophoma yunnanensis F. Liu & L. Cai, sp. nov. MycoBank MB829905. Fig. 7.
Etymology: Named after the location where it was collected from, Yunnan Province.
Sexual morph: Unknown. Asexual morph: Sterile on cultural media. Conidiomata pycnidial, on diseased leaves black, globose or subglobose, with an opening at apex, 100–200 μm diam. Pycnidial wall brown, with 3–5 layers, 13–35 μm wide, walls of textura angularis. Conidiophores reduced to conidiogenous cells lining inner cavity. Conidiogenous cells hyaline, smooth, phialidic, ampulliform or globose, aseptate, 2.5–5 × 2–4.5 μm (av. = 3.5 ± 0.5 × 3.1 ± 0.8 μm). Conidia hyaline, aseptate, granular to guttulate, ellipsoid or cylindrical, 3.5–5 × 2–3 μm (av. = 4.3 ± 0.4× 2.4 ± 0.2 μm).
Culture characteristics: On PDA, flat with entire edge, pale grey in the centre, buff at the edge, reverse blackish yellow in the centre and buff at the edge, reaching 31–34 mm diam after 10 d at 25 °C; On MEA, flat with entire edge, front and reverse blackish green, reaching 29 mm diam after 10 d at 25 °C.
Typus: China, Yunnan Province, Xishuangbanna, Mengla County, Laomansa, on Camellia sinensis, 19 Apr. 2015, F. Liu, LMS001P (holotype HMAS 248084, culture ex-type CGMCC 3.19529 = LC6753 = LF1434).
Additional material examined: China, Yunnan Province, Xishuangbanna, Mengla County, Daqishu, on Camellia sinensis, 19 Apr. 2015, F. Liu, DQS001P, living culture LC6532 = LF1167.
Notes: The leaf spots of arboreal tea plants where S. yunnanensis was isolated from were scattered, grey to brown in colour. Setophoma yunnanensis is phylogenetically related to S. yingyisheniae (Fig. 2, 96 % sequence similarity on ITS, 88 % on gapdh, 87 % on tef-1α and 93 % on tub2), but differs both morphologically and geographically from the latter (see notes under S. yingyisheniae).
DISCUSSION
Leaf spots on tea trees appeared to be caused by several previously undescribed Setophoma species. These were found on both old and young tea leaves collected from several Chinese provinces. Phylogenetic analyses provided a clear resolution for these novel Setophoma species (Figs 1, 2). Although the genus Setophoma was only recently proposed and its relatives in Pleosporales have already been partly reassessed (e.g. Aveskamp et al. 2010, de Gruyter et al. 2010, Woudenberg et al. 2013), a large number of published phoma-like names still remained untreated. Therefore, in order to avoid proposing new Setophoma names for the old published taxa, we compared our species with all known Pleosporales species associated with Camellia from China, i.e. Alternaria alternata, A. longipes, A. brassicae, Coniothyrium palmarum, Deuterophoma sp., Fusicladium theae, Hendersonia theae, Phoma camelliae, P. chinensis, P. herbarium var. herbarum, Piggotia sp., Remotididymella destructiva and Stagonosporopsis cucurbitacearum (Farr & Rossman 2019). Most of these species are clearly distinctive from Setophoma by either morphological or molecular data, but this could unfortunately not be determined for Phoma camelliae and P. chinensis, as no reliable information regarding type specimen or publication information could be found for these species. These two names were thus ignored in this study and the taxonomic treatments of P. camelliae and P. chinensis await further study.
Of the four novel species isolated from the tea plant leaf spots, three species (S. antiqua, S. longinqua and S. yunnanensis) were only isolated from old and arboreal tea plants (~100–300-yr-old, with less disease management and anthropogenic interference, Figs 3A, 7A, B) in Yunnan Province. In contrast, the fourth species, S. yingyisheniae, was widely distributed in all investigated provinces except Yunnan, and to our knowledge it only occurs on terraced tea plants (~50-yr-old), with intensive disease management and anthropogenic interference (Fig. 6A). Setophoma yingyisheniae has been isolated from both symptomatic and asymptomatic leaves tissues, indicating an alternative lifestyle ranging from endophytic to plant pathogenic. Morphologically, S. yingyisheniae differs from other Setophoma spp. in producing branched conidiophores, while other species in the genus produce solitary conidiophores. In contrast, S. endophytica was only isolated from healthy leaves, and whether it actually causes disease awaits to be determined.
A recently published species, Setophoma cyperi (Crous et al. 2016) should be excluded from the genus Setophoma, as it does not cluster in Setophoma s. str. (Fig. 1). Setophoma cyperi appears to be more closely related to Sulcispora pleurospora (Fig. 1). The definitive generic placement of S. cyperi should be clarified in future work when a broader sampling of Phaeosphaeriaceae species becomes available.
ACKNOWLEDGEMENTS
We thank Yingyi Shen, Zhifeng Zhang, Yongzhao Diao, Peng Zhao, Mengmeng Wang, Dianming Hu, Xiaoming Tan, Qian Chen, Hanxing Zhang, Zuoru He, Haoshan Fu, Xiaoyan Dao and Tianwen Hou for their help in the collection of samples. Dr William Quaedvlieg is thanked for critically revising the English text. This study was financially supported by the Project for Fundamental Research on Science and Technology, Ministry of Science and Technology of China (2014FY120100) and the Frontier Science Research Project of the Chinese Academy of Sciences (QYZDB-SSW-SMC044).
REFERENCES
- Aveskamp MM, De Gruyter J, Woudenberg JHC, et al. (2010). Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology 65: 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Pirseyedi M, Hubbard S. (1999). Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91: 964–977. [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. (2014). Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. (2016). Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gruyter J, Woudenberg JHC, Aveskamp MM, et al. (2010). Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 102: 1066–1081. [DOI] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. (2019). Fungal Databases, U.S. National Fungus Collections, ARS, USDA; https://nt.ars-grin.gov/fungaldatabases/. [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kuwabara K, Urushibara T, et al. (2012). Pink root rot of squash caused by Setophoma terrestris in Japan. Plant Disease 78: 372–375. [Google Scholar]
- Johnston-Monje D, Loewen S, Lazarovits G. (2017). Mycobiomes of tomato plants with vine decline. Canadian Journal of Plant Pathology 39: 184–200. [Google Scholar]
- Liu F, Bonthond G, Groenewald JZ, et al. (2019). Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Studies in Mycology 92: 287–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang M, Damm U, et al. (2016). Species boundaries in plant pathogenic fungi: a Colletotrichum case study. British Medical Council Evolutionary Biology 16: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Weir BS, Damm U, et al. (2015). Unravelling Colletotrichum species associated with Camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35: 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, et al. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R, Liu JK, Manamgoda DS, et al. (2014). The sexual state of Setophoma. Phytotaxa 176: 260–269. [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin HD, et al. (2013). Sizing up Septoria. Studies in Mycology 75: 307–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Rehner SA, Samuels GJ. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634. [Google Scholar]
- Rivedal HM, Stone AG, Johnson KB. (2018). First report of Setophoma terrestris causing pink root rot of winter squash (Cucurbita maxima) in Oregon. Plant Disease 102: 2661. [Google Scholar]
- Thambugala KM, Wanasinghe DN, Phillips AJL, et al. (2017). Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Myco-sphere 8: 697–796. [Google Scholar]
- Vilgalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innes MA, Gelfand DH, Sninsky JJ, et al, eds). Academic Press, USA: 315–322. [Google Scholar]
- Woudenberg JHC, Groenewald JZ, Binder M, et al. (2013). Alternaria redefined. Studies in Mycology 75: 171–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zuzak K, Harding M, et al. (2017). First report of pink root rot caused by Setophoma (Pyrenochaeta) terrestria on canola. Canadian Journal of Plant Pathology 39: 354–360. [Google Scholar]