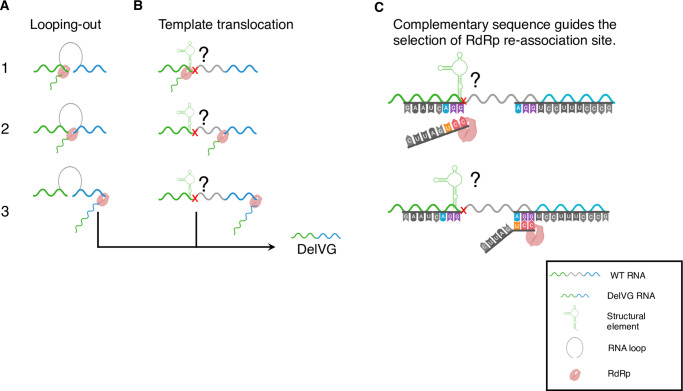
Fig 1. Proposed DelVG formation mechanisms for influenza viruses.
(A) Looping-out model. The 2 sites of the DelVG junction are brought together by an RNA loop structure (1), potentially facilitated by the hairpin structure of the viral RNP, creating a connection point where the viral polymerase complex can “roll-over” (2) resulting in deleting the loop sequence (3). (B) Template translocation model. Pausing of daughter strand synthesis by the RdRp (1) is suggested to be potentially caused by a local RNA structure in the template or basepair mismatch at the junction site (the red x). RdRP remains associated with nascent daughter strand and continues processing along the template molecule (2). At a downstream point, the RdRp resumes templated elongation of the daughter strand (3), resulting in deletion. (C) A schematic based on model B showing how complementarity within overlapping sequences at the DelVG junction sites may allow the nascent strand to re-associate with the template, where the RdRp can resume elongation. Figures generated with the assistance of Biorender.com. DelVG, deletion-containing viral genome; RdRp, RNA-dependent-RNA-polymerase; WT, wild type