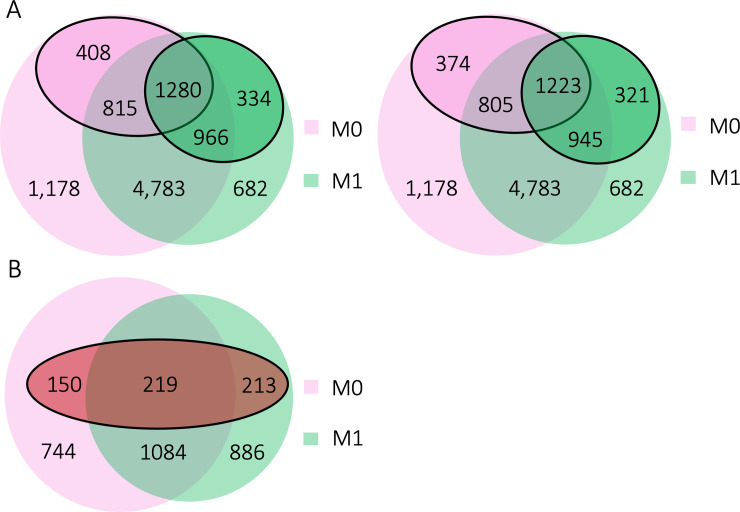
Fig 5. Genes analyzed for ASE and differential ASE analysis in the macrophage RNA-seq dataset.
(A) Total number of genes analyzed, and number of significant ASE genes in M0 (pink) and M1 (green) macrophages obtained from one-condition analysis. Solid circles indicate the nominal significant (left) and FDR-adjusted significant (right) ASE genes detected under each condition. (B) Total number of genes analyzed for two-condition analysis. Genes were selected from significant (nominal) ASE genes for M0 (pink) and M1 (green) macrophages that expressed under both conditions. Genes with less than three matched reads, i.e., the tSNP has read counts for both M0 and M1 macrophages of the same individual, were further excluded from the analysis. Solid circle indicates the FDR-adjusted significant differential ASE genes detected between M0 and M1.