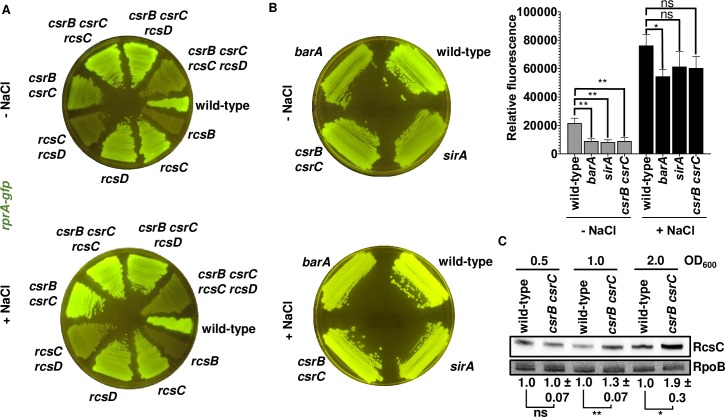
Fig 5. BarA activation of RcsB via the SirA-activated regulatory RNAs CsrB and CsrC.
(A) The regulatory RNAs CsrB and CsrC are required for full RcsB activation on LB agar plates. Fluorescence from wild-type (14028s), rcsB (EG12925), rcsC (HS1350), rcsD (HS1382), rcsC rcsD (HS1383), csrB csrC (HS1651), csrB csrC rcsC (HS1654), csrB csrC rcsD (HS1655) and csrB csrC rcsC rcsD (HS1656) Salmonella harboring plasmid pRprA-GFP (rprA-gfp) following 24 h of growth on LB solid medium without (-NaCl) or with (+NaCl) NaCl. Data are representative of two independent experiments, which gave similar results. (B) Fluorescence from wild-type (14028s), barA (HS1520), sirA (HS1565) and csrB csrC (HS1651) Salmonella harboring plasmid pRprA-GFP (rprA-gfp) following 24 h of growth on LB solid medium without (-NaCl) or with (+NaCl) NaCl. Data are representative of three independent experiments, which gave similar results. Quantification of the fluorescence is provided next to the plate images. Values derived from three independent experiments (mean ± standard deviation) were statistically analyzed by Prism 8 using two-tailed unpaired t test. Statistical significance is indicated by * P<0.05, **P<0.01; ns, not significant. Error bars indicate standard deviation. (C) Western blot analysis of crude extracts from rcsC-3XFLAG (HS539) and rcsC-3XFLAG csrB csrC (HS2263) strains grown in LB NaCl-free broth. Samples were analyzed with antibodies directed to the FLAG epitope or the RpoB protein. Data are representative of three independent experiments, which gave similar results. RcsC levels for csrB csrC mutant strain relative to wild-type Salmonella are marked below (fold increase ± standard deviation). Values were statistically analyzed by Prism 8 using two-tailed unpaired t test. Statistical significance is indicated by * P<0.05, ** P<0.01; ns, not significant.