Figure 2. Neurons in nat.
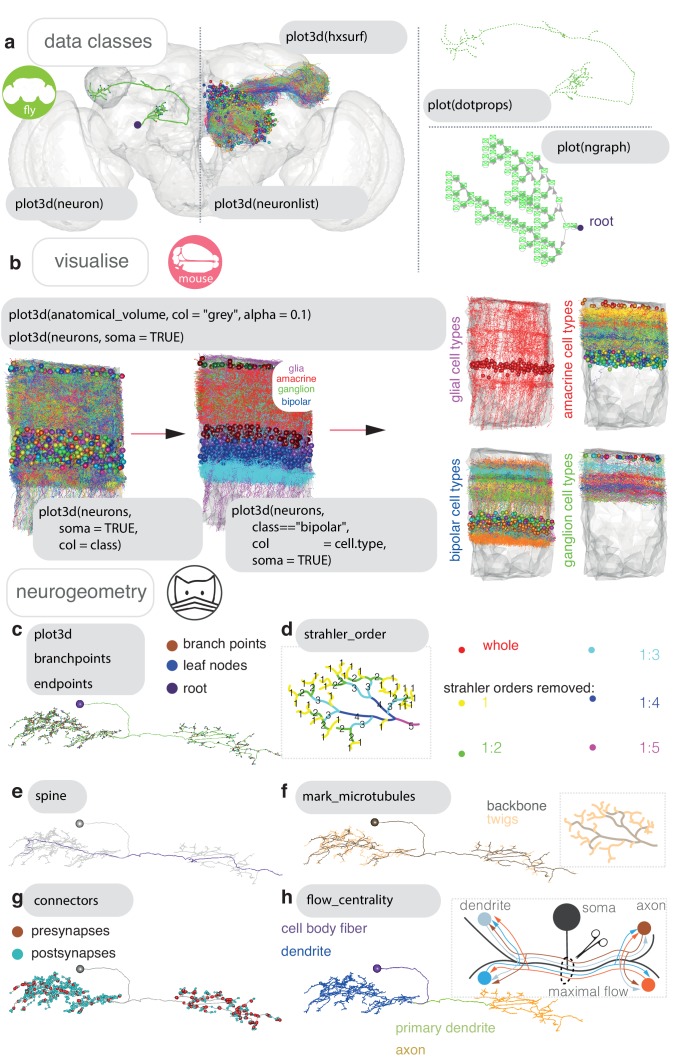
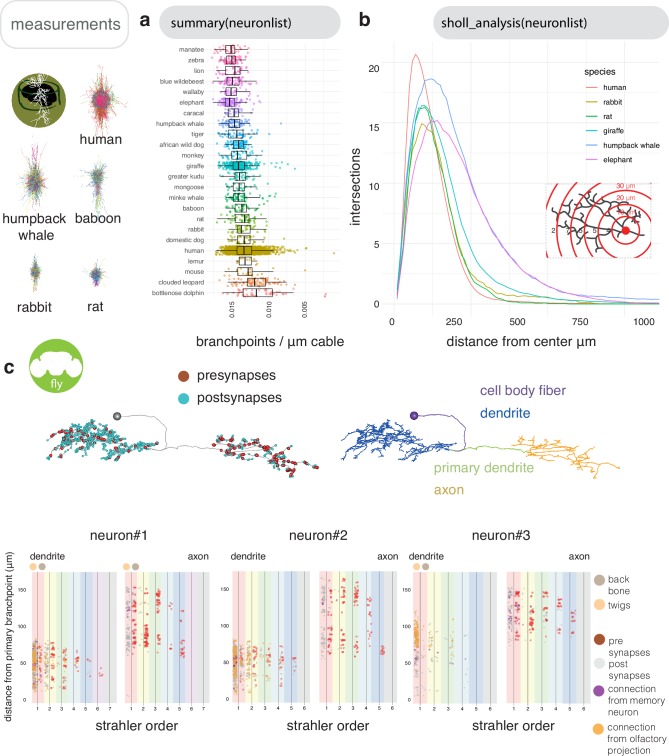
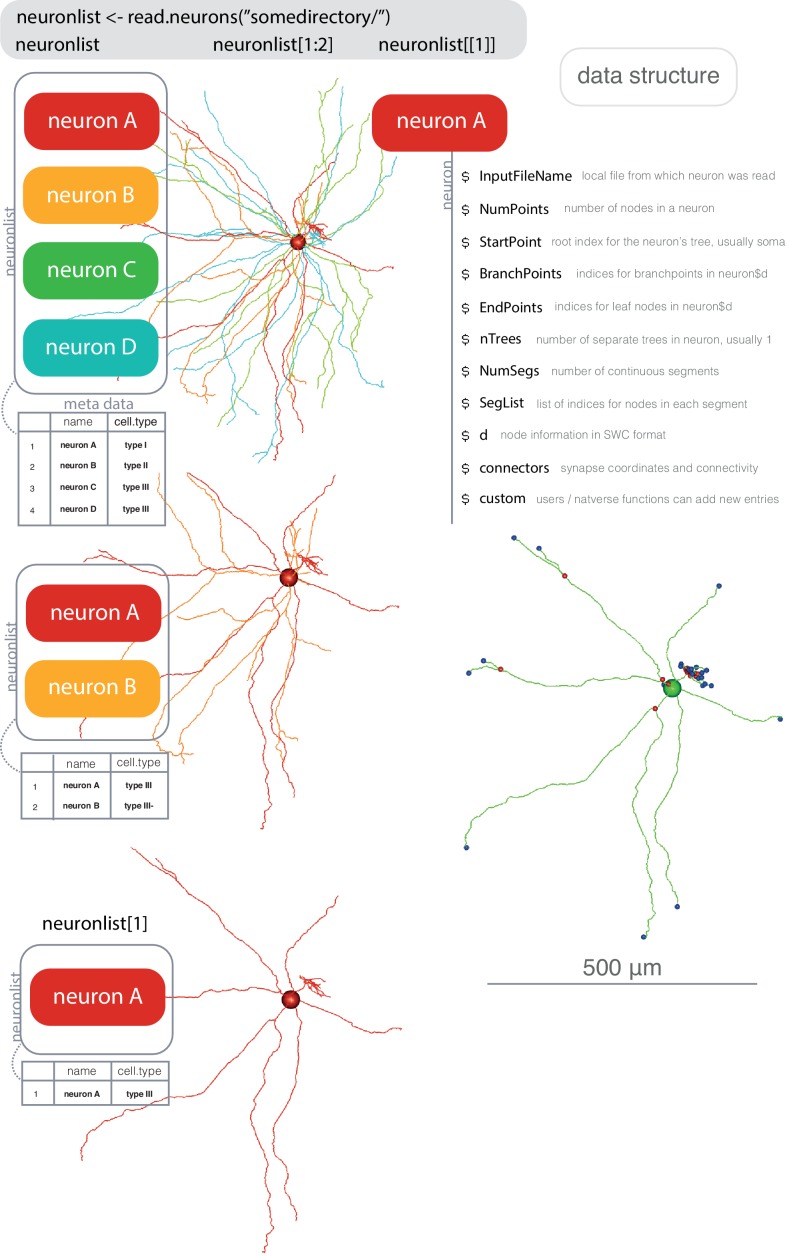
(a) Data classes defined by nat. A D. melanogaster DA1 olfactory projection neuron (Costa et al., 2016) is shown as part of four different data types, useful for different sorts of analyses: as a neuron object (left), as part of a larger neuronlist of multiple olfactory projection neurons (middle), as a vector-cloud dotprops object (right, upper) and a ngraph object (right, lower). In grey, the FCWB template brain, a hxsurf object from the package nat.flybrains, is shown. Generic code for visualizing these data types shown. (b) Visualisation, with generic sample code, of connectomic data from a dense reconstruction inner plexiform layer of the mouse retina is shown, coloured by the cell class and then cell type annotations given by their reconstructors (Helmstaedter et al., 2013). Because this dataset contains many neuron fragments that have been severely transected, we only consider skeletons of a total cable length greater than 500 μm using functions summary and subset. Somata are shown as spheres. (c) A synaptic-resolution neuron reconstruction for a D. melanogaster lateral horn neuron (Dolan et al., 2018a) has been read from a live CATMAID project hosted by Virtual Fly Brain (https://fafb.catmaid.virtualflybrain.org/) using catmaid, and plotted as a neuron object. It is rooted at the soma, consistent with the convention. (d) Boxed, Strahler order is a measure of branching complexity for which high Strahler order branches are more central to a neuron’s tree structure, and the lower order ones more peripheral, such that branches with leaf nodes are all Strahler order 1. Main, the same neuron which has had its lower Strahler order branches (see inset) progressively pruned away. (e) We can extract the longest path through a neuron, its ‘spine’, purple, a method that could help define the tracts that a neuron might traverse. (f) Boxed, in insect neurons, the main structure of the neuron is supported by a microtubular backbone. As it branches its more tortuous, smaller caliber neurites loose the microtubule, and make more postsynapses (Schneider-Mizell et al., 2016). Main, in CATMAID users can tagged the tree nodes that mark the position where neurite loses its microtubular backbone, so an R user can use prune family functions to remove, add or differentially colour microtubular backbone versus twigs. (g) Both presynapses and postsynapses can been manually annotated in CATMAID, and be visualised in R. Because neurons from CATMAID have synaptic data, they are treated as a slightly different class by the natverse, called catmaidneuron. A neuronlist can also comprise many catmaidneuron objects. (h) Right, using synaptic information, it is possible to use a graph theoretic approach which divides the neuron at the point of maximum ‘flow’ - the region in the neuron at which there are the most parallel paths - having ‘drawn’ a path between each input synapse and each output synapse that pass through every node on the skeleton (Schneider-Mizell et al., 2016). This helps divide a neuron into its dendrites, axon, intervening cable (maximum flow, the primary dendrite) and its cell body fiber (no flow). In insects, the cell body lies outside the neuropil and is connected to its arbour by a single fiber. Main, axon-dendrite split shown for exemplary neuron using seesplit3d.