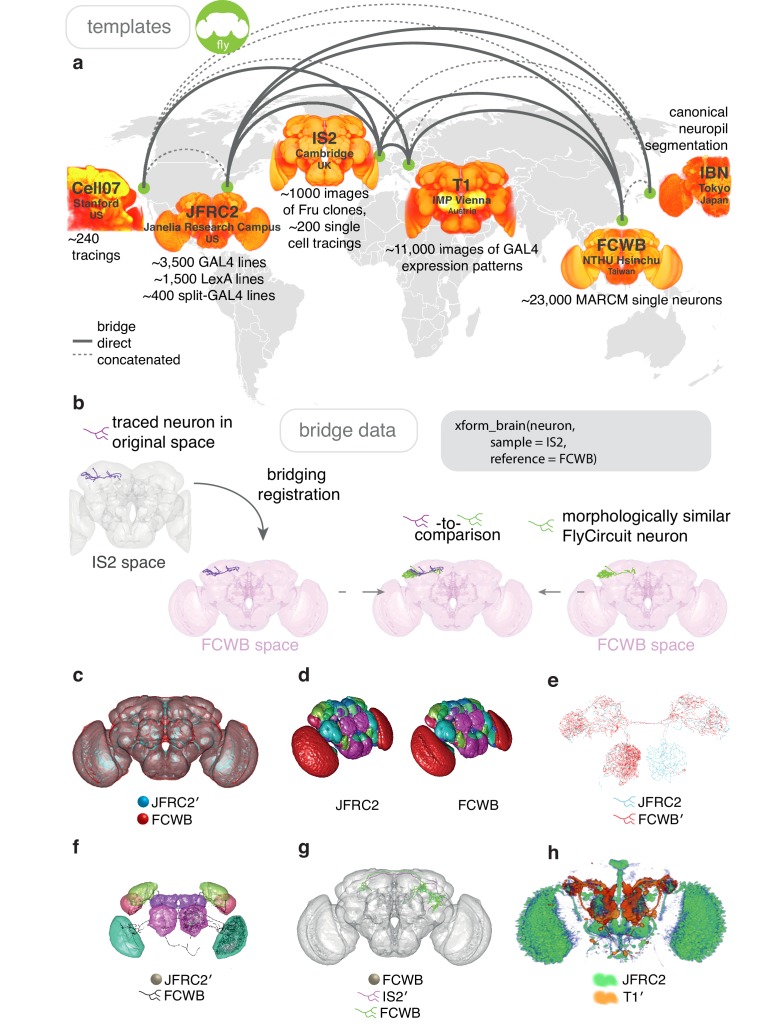
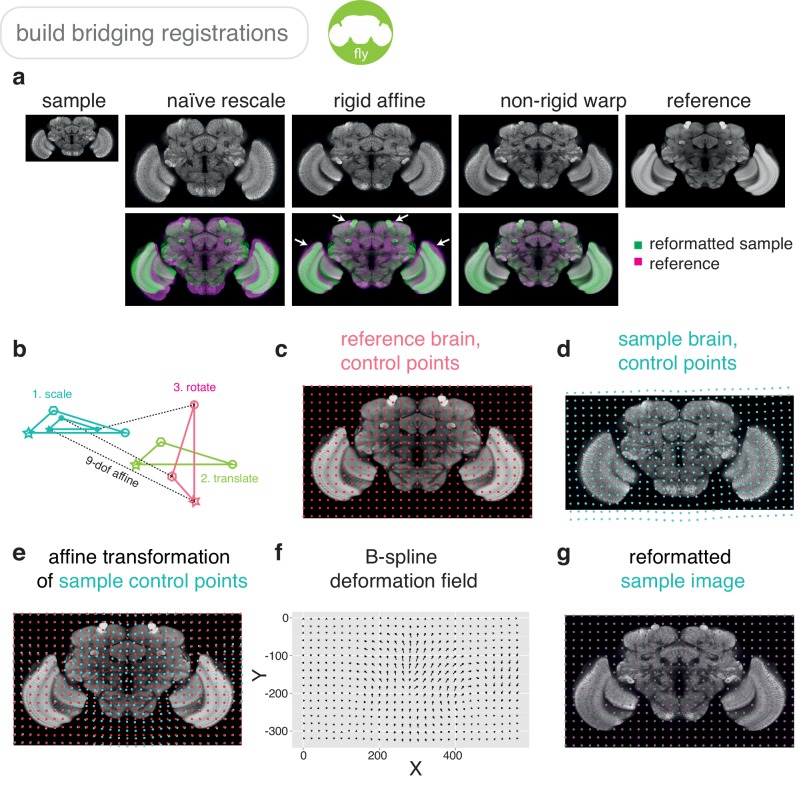
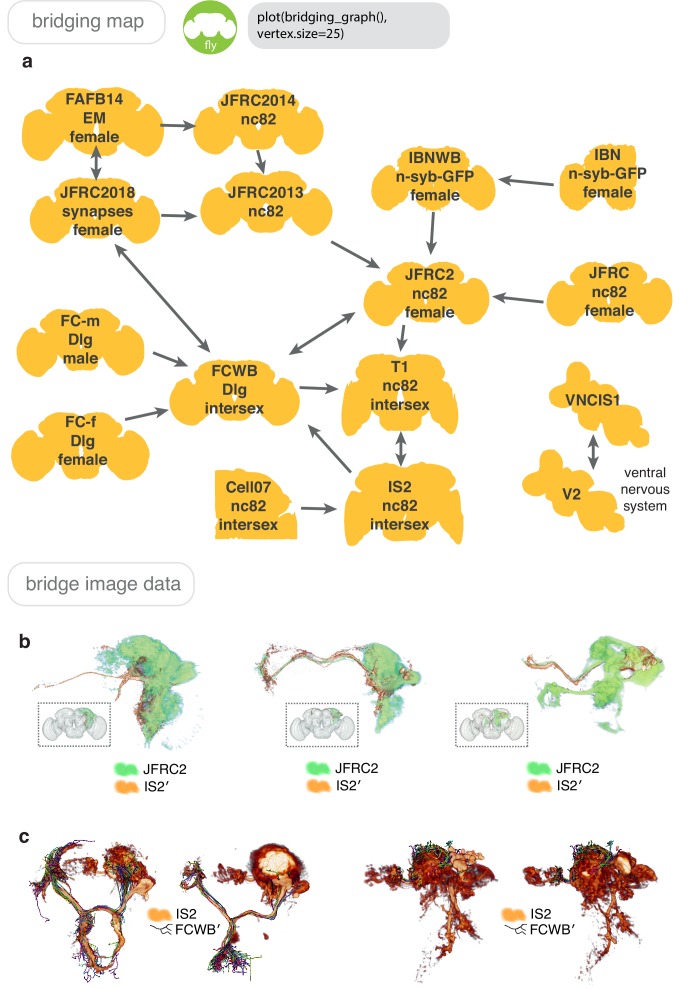
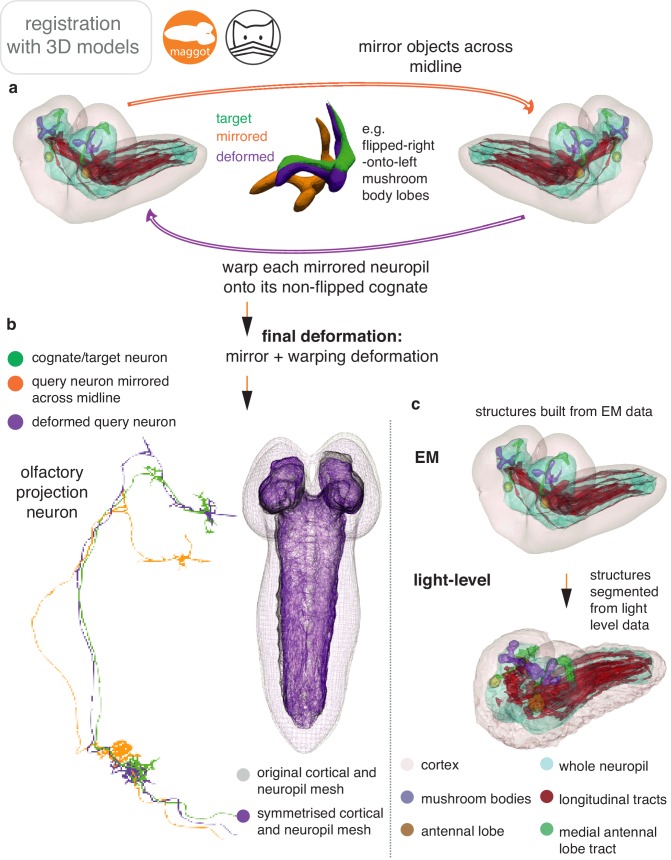
Figure 6. Bridging registrations for brain templates.
(a) A small sample of Drosophila template brains used around the world are shown. (b) A partial neuron tracing (purple) made using Simple Neurite Tracer (Longair et al., 2011) being transformed (xform_brain) from the IS2 standard brainspace to the FCWB, where it can be co-visualised with a more complete, matching morphology from FlyCircuit, using our R package flycircuit. (c) Outermost neuropil boundaries for FlyCircuit (red) and FlyLight (cyan) template brains. Primed space names indicate original spaces for data. Unprimed space names indicate current space of image. (d) Neuropil segmentation from JFRC2 space alongside FCWB reformatted version. (e) CSD interneuron from FlyCircuit (red) and FlyLight, GMR GAL4 expression pattern (cyan). (f) Neuropil segmentation from JFRC2 (Ito et al., 2014) space that has been bridged into FCWB space, so it can be seen along with selected neurons from FlyCircuit. (g) A traced neuron in FCWB space alongside morphologically similar neuron from FlyCircuit. (h) Expression pattern of Vienna Tiles line superimposed on expression pattern of FlyLight line. (Since we made our bridging publically available in April 2014, Otsuna et al., 2018 have also, separately, bridged these two datasets.).