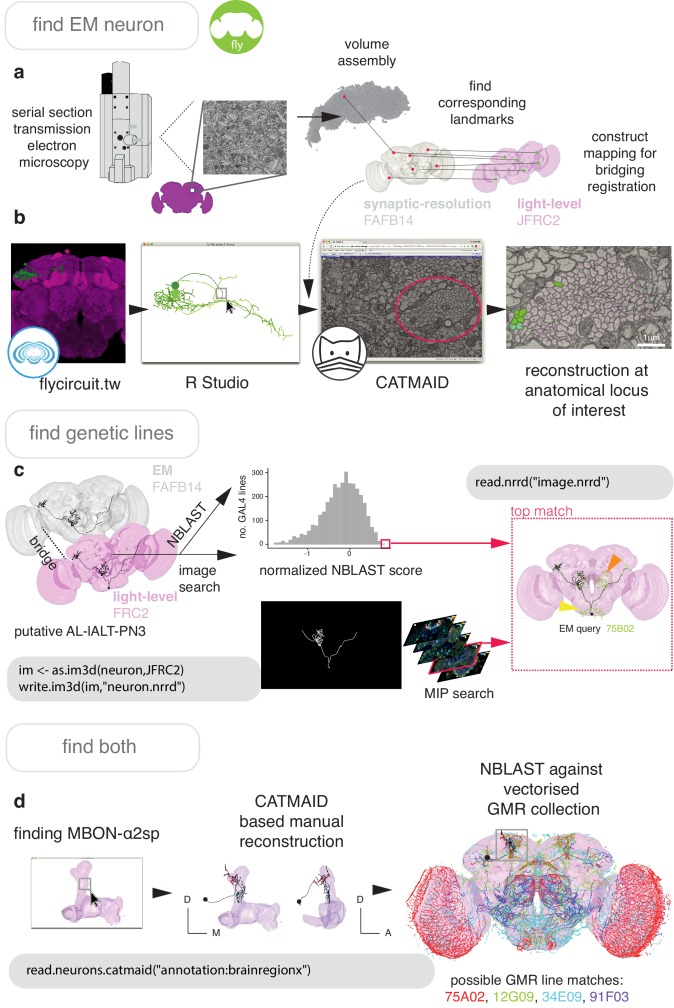
Figure 7. Finding specific neurons in EM and LM data.
(a) Pipeline for acquiring EM neuron data. Serial section transmission EM at high speed with a TEM camera array (Bock et al., 2011) produced several micrographs per section at 4 × 4 nm resolution, ~40 nm thick. These were, per section, stitched into mosaics which were, across sections, registered to create the female adult fly brain v.14 template space (FAFB14, grey) (Zheng et al., 2018). Corresponding landmarks between FAFB14 and JFRC2 can be found and used to build a bridge. (b) The R package elmr can be used to select an anatomical locus, here the PD2 primary neurite tract (Frechter et al., 2019), from 3D plotted light-level neurons, taken from FlyCircuit, and generate a URL that specifies its correct coordinates in a FAFB14 CATMAID instance. Candidates (185) may then be coarsely traced out until they deviate from the expected light-level morphologies (178 pink dotted profiles, often a few minutes to an hour of manual reconstruction time to rule out neurons of dissimilar cell types sharing a given tract, similar cell types are more subtly different and might need to be near completely reconstructed). Those that remain largely consistent were fully reconstructed (green profiles, ~7–12 person-hours per neuron) (Li et al., 2019). (c) Close matches reveal likely morphology of non-reconstructed branches (orange arrow) but also contain off-target expression (yellow arrow). Identification of multiple candidate lines enables split-GAL4 line generation aimed at retaining common neurons in two GAL4 patterns. MultiColor FlpOut (MCFO) (Nern et al., 2015) of resultant splits can be compared with the EM morphology. Here, a candidate GAL4 line is found for AL-lALT-PN3 (Frechter et al., 2019; Tanaka et al., 2012) using NBLAST and a MIP search (Otsuna et al., 2018). (d) A recent dense, but volume-restricted reconstruction of the mushroom body α-lobe discovered a ‘new’ mushroom body output neuron type (MBON-α2sp) (Takemura et al., 2017). By bridging from the correct mushroom-body compartment using a mushroom body mesh (Ito et al., 2014) visualised in R Studio, to the FAFB14 EM data’s equivalent space in CATMAID using the R package elmr, an experienced tracer can easily identify dendrites and find MBON-α2sp. By doing so, we found its previously unreported axon-morphology. We then imported the skeleton into R studio, bridged MBON-α2sp into the JFRC2 template space where it could be NBLAST-ed against GMR GAL4 lines to identify candidate lines containing the MBON.