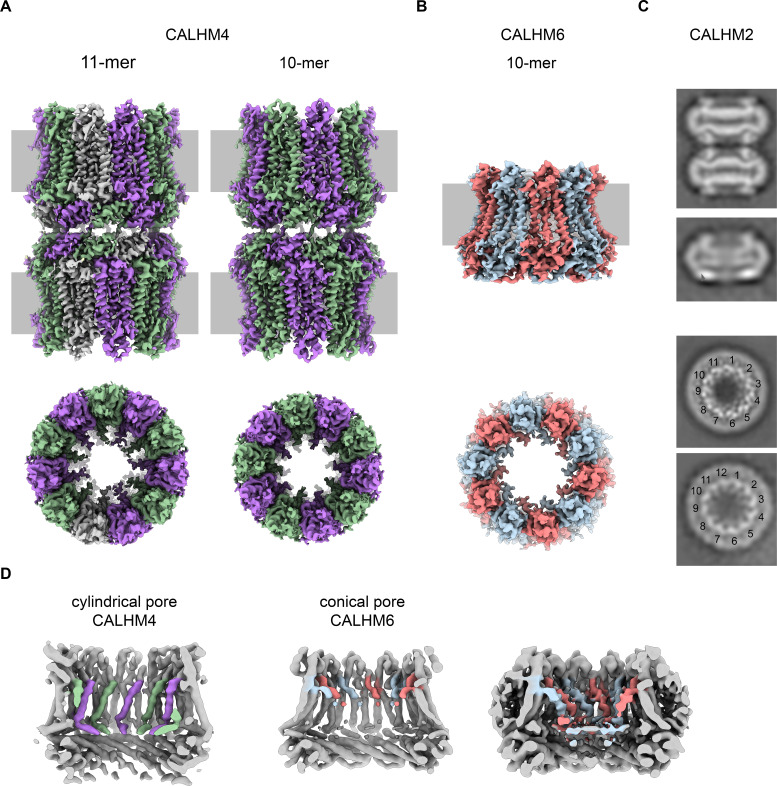
Figure 3. Cryo-EM analysis.
(A) Cryo-EM density of undecameric (11-mer) and decameric (10-mer) pairs of CALHM4 channels at 3.8 and 4.1 Å respectively. Data were recorded from a Ca2+-free sample. Subunits are colored in lilac and green, respectively. (B) Cryo-EM density of decameric CALHM6 channels at 4.4 Å. Subunits are colored in red and light-blue, respectively. A, B, Views are from within the membrane with membrane indicated as grey rectangle (top) and from the outside (bottom). (C) Selected 2D classes of the CALHM2 data showing interacting channel pairs and single channels viewed from within the membrane (top) and views of undecameric and dodecameric channels with subunits numbered (bottom). (D) Slices through the CALHM4 (left) and the CALHM6 (right) channels illustrating the distinct features of the cylindrical and conical pore conformations. View of CALHM6 at lower contour (right) shows extended density for the mobile TM1. Maps are low-pass filtered at 6 Å. Colored features refer to density corresponding to TM1 and NH.