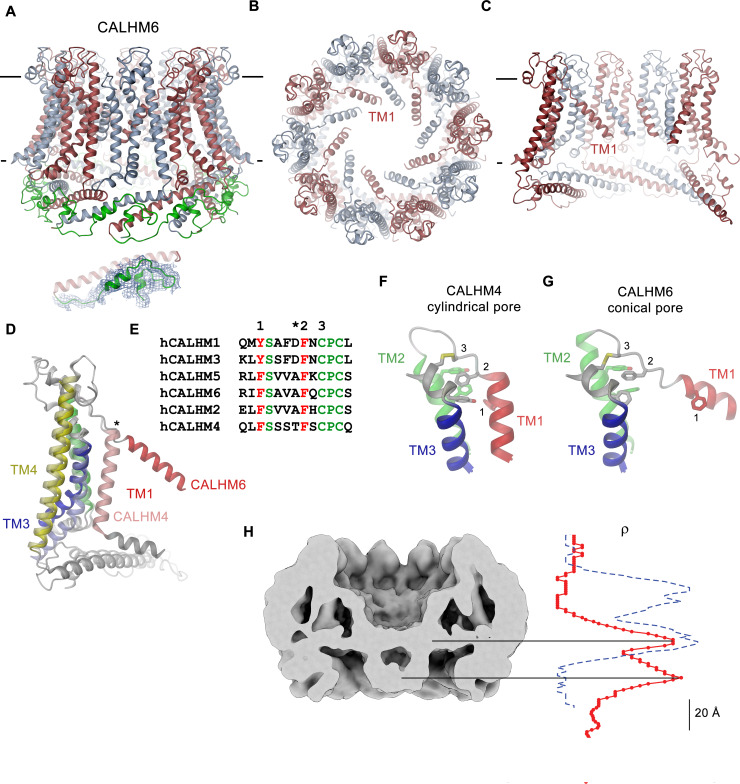
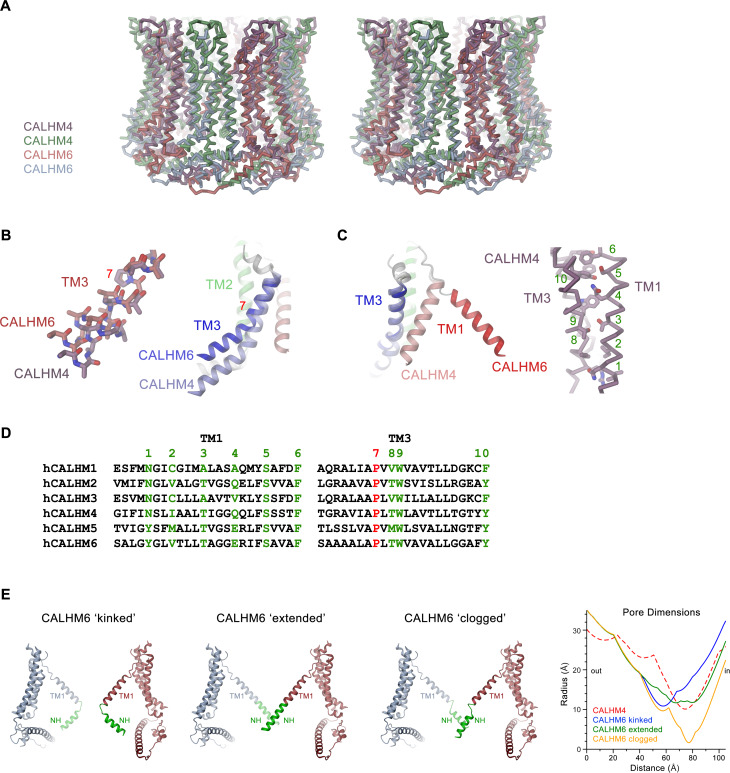
Figure 7. CALHM6 structure.
(A) Ribbon representation of the decameric CALHM6 structure viewed from within the membrane. Subunits are colored in red and light blue. The C-terminus following CTH is colored in green. Inset (below) shows a close-up of this region with cryo-EM density superimposed. (B) View of the CALHM structure from the extracellular side. (C) Slice through the pore of the CALHM6 structure. B, C, TM1, which has moved towards the pore axis is labeled. A, C, the membrane boundary is indicated. (D) Superposition of single subunits of the CALHM4 and CALHM6 structures illustrating conformational changes. Coloring is as in Figure 5A with CALHM4 shown in brighter shades of the same color. Secondary structure elements are labeled and the hinge for the movement of TM1 is indicated by an asterisk. (E) Sequence alignment of the end of TM1 and the following loop of CALHM paralogs. Selected conserved residues are colored in green and red. Numbering corresponds to residues indicated in panels F and G. Asterisk marks the hinge region displayed in D. Close-up of the extracellular region involved in conformational changes in (F), the cylindrical conformation displayed in CALHM4 and (G), the conical conformation displayed in CALHM6. Residues contributing to a cluster of aromatic residues on TM1-3 and a conserved disulfide bridge are shown as sticks. Selected positions highlighted in E are labeled. F, G, Coloring is as in Figure 5A. (H) Slice through the pore region of the CALHM6 decamer viewed from within the membrane. Shown is non-averaged density at low contour to highlight the location of diffuse density within the pore. A plot of the density along the pore axis of CALHM6 is shown in red, the corresponding density in CALHM4 is shown as a dashed blue line for comparison. The two maxima in the CALHM6 density are shifted towards the intracellular side. The density corresponding to the headgroups of the outer leaflet of the bilayer in CALHM4 is absent. Density at the location of the headgroup region at the inner leaflet of the bilayer and further towards the intracellular side could correspond to either lipids or to the poorly ordered N-terminus.