Abstract
Changes in environmental temperature influence cellular processes and their dynamics, and thus affect the life cycle of organisms that are unable to control their cell/body temperature. Meiotic recombination is the cellular process essential for producing healthy haploid gametes by providing physical links (chiasmata) between homologous chromosomes to guide their accurate segregation. Additionally, meiotic recombination—initiated by programmed DNA double-strand breaks (DSBs)—can generate genetic diversity and, therefore, is a driving force of evolution. Environmental temperature influencing meiotic recombination outcome thus may be a crucial determinant of reproductive success and genetic diversity. Indeed, meiotic recombination frequency in fungi, plants and invertebrates changes with temperature. In most organisms, these temperature-induced changes in meiotic recombination seem to be mediated through the meiosis-specific chromosome axis organization, the synaptonemal complex in particular. The fission yeast Schizosaccharomyces pombe does not possess a synaptonemal complex. Thus, we tested how environmental temperature modulates meiotic recombination frequency in the absence of a fully-fledged synaptonemal complex. We show that intragenic recombination (gene conversion) positively correlates with temperature within a certain range, especially at meiotic recombination hotspots. In contrast, crossover recombination, which manifests itself as chiasmata, is less affected. Based on our observations, we suggest that, in addition to changes in DSB frequency, DSB processing could be another temperature-sensitive step causing temperature-induced recombination rate alterations.
Electronic supplementary material
The online version of this article (10.1007/s10577-020-09632-3) contains supplementary material, which is available to authorized users.
Keywords: meiosis, meiotic recombination, environmental temperature, Schizosaccharomyces pombe
Introduction
Single-celled organisms, such as yeasts, and other organisms unable to control their internal temperature are at the mercy of the environmental temperature, which can fluctuate considerably between seasons and during day-night cycles. Schizosaccharomyces pombe and Saccharomyces cerevisiae are globally distributed and very distantly related species with a rather poorly understood ecology (Liti 2015; Jeffares 2018), but it is likely that they are exposed to changing temperatures in their respective niches. Changes in temperature can modulate meiotic recombination outcome in a range of organisms, including fungi (Plough 1917; Lu 1974; Rose and Baillie 1979; Börner et al. 2004; Pryce et al. 2005; Higgins et al. 2012; Phillips et al. 2015; Zhang et al. 2017; Lloyd et al. 2018; Modliszewski et al. 2018). Accordingly, environmental temperature has been suggested to be a major driver in the evolutionary adaptation of the meiotic machinery (Bomblies et al. 2015).
The main function of meiotic recombination is to ensure correct chromosome segregation, as it establishes physical connections between the homologous chromosomes (homologues). This is achieved through the repair of programmed DNA double-strand breaks (DSBs) from the homologue rather than the sister chromatid, and the processing of recombination intermediates between the homologues as crossovers (COs) (de Massy 2013; Hunter 2015). The transesterase Spo11 makes the DSBs, preferentially in particular regions called hotspots (de Massy 2013; Tock and Henderson 2018). Subsequently, the DSB ends are resected to enable homologous recombination, which ultimately leads to COs and non-crossovers (NCOs) depending on the repair pathway (de Massy 2013; Gray and Cohen 2016).
Meiotic recombination is influenced by the chromatin environment and in most organisms by the formation of a meiosis-specific axis along the chromosomes. These axes are then connected by filamentous proteins to build the synaptonemal complex (Hunter 2015; Gray and Cohen 2016). Proteins forming the chromosome axis and/or the synaptonemal complex are important to enable meiotic recombination or at least to maintain it at wild-type levels (de Massy 2013; Hunter 2015; Gray and Cohen 2016). A large part of the effect on meiotic recombination frequency or event placement exerted by temperature changes seems to be mediated by the chromosome axis and the synaptonemal complex in many organisms (Morgan et al. 2017). Fission yeast lacks a canonical synaptonemal complex (Olson et al. 1978; Bähler et al. 1993; Molnar et al. 2003). It only forms the so-called linear elements, which are meiotic chromosome axes evolutionarily related to the lateral elements of the synaptonemal complex (Lorenz et al. 2004; Loidl 2006, 2016). Sz. pombe is, thus, an ideal system to test the response of recombination to temperature changes in the absence of a fully-fledged synaptonemal complex.
Here, we employed a series of genetic and cytological assays to test whether fission yeast meiosis and meiotic recombination are susceptible to temperature changes. We determined the full ‘fertile range’ (Bomblies et al. 2015) of the Sz. pombe laboratory strain, and measured meiotic intra- and intergenic recombination frequencies at and around ade6 using a genetic assay (Lorenz et al. 2010). We find that intergenic recombination around ade6 is not strongly affected over the fertile range in Sz. pombe, but that gene conversion, especially at hotspots engendered by point mutants in ade6, is cold-sensitive. Further experimentation indicates that in addition to changes in the formation of DSBs (Hyppa et al. 2014), DSB processing could be another source for a reduced gene conversion frequency particularly at temperatures below 25 °C.
Material and methods
Yeast strains and culture conditions
Cells were cultured on yeast extract (YE), and on yeast nitrogen base glutamate (YNG) agar plates containing the required supplements (concentration 250 mg/l on YE, 75 mg/l on YNG). Crosses were performed on malt extract (ME) agar containing supplements at a final concentration of 50 mg/l (Sabatinos and Forsburg 2010).
All Schizosaccharomyces pombe strains used for this study were either published previously, or have been generated from existing strains by crossing (see Table S1). Different ade6 alleles (Table S2) were introduced by crossing the respective mutant ade6 strain with ade6+ strains carrying the ura4+ and his3+ artificially introduced markers (aim) (UoA95, UoA96, UoA97, UoA98) (Osman et al. 2003). The point mutations in the ade6 alleles were verified by Sanger DNA sequencing (Source BioScience, Nottingham, UK) (Table S2).
Epitope tagging of hop1+ with a C-terminal 13myc-kanMX6 tag has been described in detail (Brown et al. 2018).
Genetic and cytological assays
Determination of spore viability by random spore analysis and the meiotic recombination assay were performed as previously described (Osman et al. 2003; Sabatinos and Forsburg 2010).
Meiotic time-courses and preparation of chromatin spreads were in essence performed as published (Loidl and Lorenz 2009), except for the use of 100 mg/ml Lallzyme MMX (Lallemand Inc., Montréal, Canada) as the cell-wall digesting enzyme (Flor-Parra et al. 2014). Immunostaining was performed according to an established protocol (Loidl and Lorenz 2009) using polyclonal rabbit α-myc (ab9106; Abcam PLC, Cambridge, UK) at a 1:500 dilution and monoclonal rat α-GFP [3H9] (ChromoTek GmbH, Planegg-Martinsried, Germany) at a 1:100 dilution as primary antibodies. Antibody-bound protein was visualized using donkey α-rabbit IgG AlexaFluor-555 (ab150062; Abcam) and donkey α-rat IgG AlexaFluor-488 (ab150153; Abcam), both at a 1:500 dilution. DNA was stained by Hoechst 33342 (Molecular Probes, Eugene, OR, USA) at a final concentration of 1 μg/ml.
Black-and-white images were taken with a Zeiss AxioCam MRm CCD camera (controlled by AxioVision 40 software v4.8.2.0) mounted on a Zeiss Axio Imager.M2 (Carl Zeiss AG, Oberkochen, Germany) epifluorescence microscope equipped with the appropriate filter sets to detect red, green and blue fluorescence. Individual images were acquired for each channel to detect Hop1-13myc, Rec7-GFP, Rad11-GFP and Hoechst 33342. Images were pseudo-coloured and overlayed using Adobe Photoshop CC (Adobe Systems Inc., San José, CA, USA). Immunodetected Rec7-GFP and Rad11-GFP foci were counted on images of meiotic prophase I nuclei at the thread and network stages identified by the presence of immunostained Hop1-13myc linear elements (Lorenz et al. 2006) within the Hoechst 33342-positive area using the ‘count’ function in Adobe Photoshop CC.
Data presentation and statistics
Raw data is available on figshare (10.6084/m9.figshare.11192861). Line graphs and bar charts were produced using Microsoft Excel 2016 (version 16.0.4638.1000, 32-bit), and scatter plots were generated in GraphPad Prism 5 for Windows (version 5.04). Box-and-whisker plots were created in R (version i386, 3.0.1) (http://www.r-project.org/) (Lorenz et al. 2014). R was also used to compute Kruskal-Wallis test and Tukey’s Honest Significant Differences employing the kruskal.test() and TukeyHSD() functions, respectively. Mann-Whitney U tests were performed as previously described (Lorenz et al. 2014).
Results
The fertile range of fission yeast lies between 11 and 33 °C
Bomblies and co-workers recently noted that in order to understand temperature effects on meiotic recombination, it is important to know the ‘fertile range’ of a species; otherwise, the results will be skewed by including temperatures outside or omitting temperatures within the fertile range (Bomblies et al. 2015; Lloyd et al. 2018). We set up matings of prototrophic fission yeast strains (ALP714 × ALP688) in a temperature range between + 4 and + 35 °C on sporulation media. Matings were checked regularly whether asci containing spores were observed within 30 days. No asci were observed at + 4 and at + 35 °C after 1 month of incubation, putting the fertile range of Sz. pombe somewhere between these two temperatures. Indeed, mating at 11 °C resulted in the formation of asci containing viable spores within 14 days, at 16 °C within 7 days, at 20 °C within 5 days, at 25 and 30 °C within 3 days and at 33 °C within 2 days (Fig. 1).
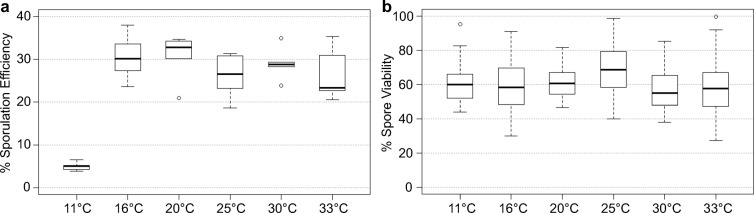
Fig. 1.
The fertile range of Schizosaccharomyces pombe. The upper, middle and lower lines of the box represent the third, second and first quartile, respectively (second quartile = median). The ‘whiskers’ represent the minimum and maximum of the range, unless they differ more than 1.5-times the interquartile distance from the median, then the 1.5-times interquartile distance around the median is indicated by the ‘whiskers’, and outliers are shown as open circles. a Sporulation efficiency in % determined in crosses of ALP714 × ALP688 at 11 °C after 14d (n = 7), at 16 °C after 7d (n = 6), at 20 °C after 5d (n = 5), at 25 °C after 3d (n = 6), at 30 °C after 2d (n = 6) and at 33 °C after 2d (n = 6). b Cumulative spore viability in % encompassing all data in Fig. 3 at 11 °C after 14d (n = 11), at 16 °C after 7d (n = 64), at 20 °C after 5d (n = 46), at 25 °C after 3d (n = 75), at 30 °C after 2d (n = 48) and at 33 °C after 2d (n = 59). For details of data see Tables S3 for (a) and S4 for (b)
Sporulation efficiency (i.e. the percentage of asci containing spores among the total population of cells) was ~ 25% at all temperatures, except at 11 °C when it was only ~ 5% (Fig. 1a, Table S3).
We also monitored spore viability by random spore analysis (Fig. 1b) during the following meiotic recombination assays (Figs. 2 and 3) performed at 11, 16, 20, 25, 30 and 33 °C (within the fertile range). At all temperatures tested, spore viability was ~ 60% (Fig. 1b), indicating that even at 11 °C when sporulation was rather inefficient (Fig. 1a), the viability of the spores that did develop was normal.
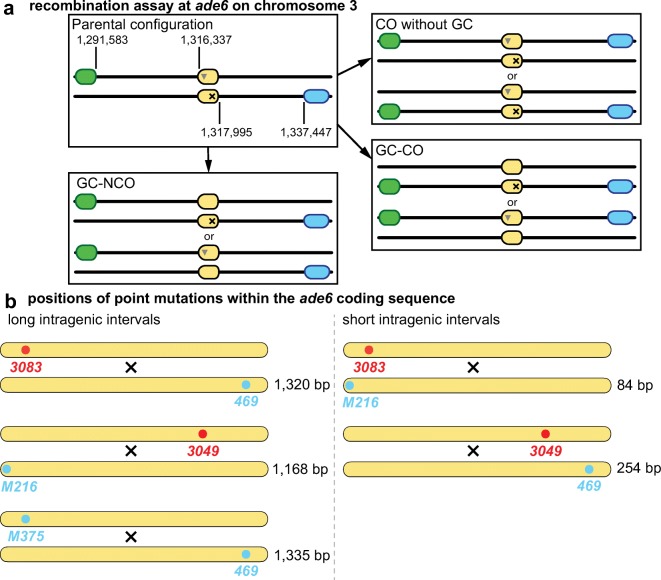
Fig. 2.
Meiotic recombination assay composed of different ade6 alleles flanked by artificially introduced markers ura4+-aim2 & his3+-aim. a Schematic showing the meiotic recombination assay at ade6 (yellow) and its common outcomes. Ade+ recombinants arise via gene conversion (GC) associated with a crossover (GC-CO) or a non-crossover (GC-NCO). The positions of ade6 and the artificially introduced markers ura4+-aim2 (green) and his3+-aim (light blue) on chromosome 3 are indicated [in bps]. Positions of point mutations are shown as ▼ and ×. b Schematic of the ade6 coding sequence indicating the point mutations and their positions (approximately to scale) used in the recombination assays, hotspots are indicated in red and non-hotspots in light blue. The distance between the sequence polymorphisms across the homologues is indicated in relation to the given hotspot of each cross [in bp]
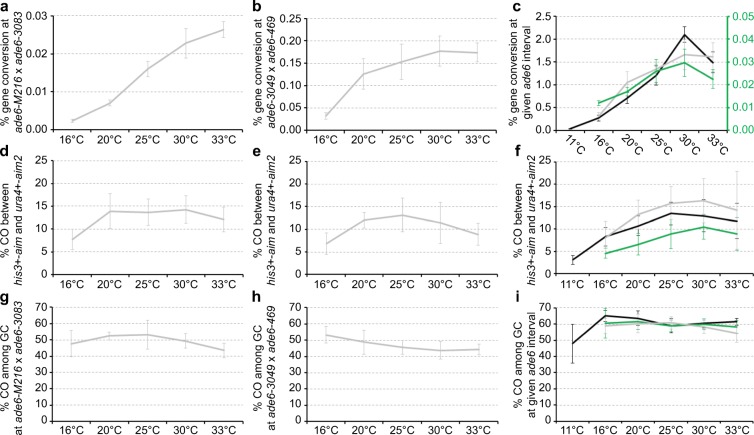
Fig. 3.
Environmental temperature alters recombination frequency at ade6. Frequency of a–c gene conversion in wild type at the indicated intragenic ade6 interval, d–f crossover (CO) between his3+-aim and ura4+-aim2 and g–i CO between his3+-aim and ura4+-aim2 among gene conversion (GC) events at ade6 from crosses performed at different temperatures. (a, d, g) UoA110 × UoA100: 16 °C (n = 15), 20 °C (n = 10), 25 °C (n = 12), 30 °C (n = 12), 33 °C (n = 12). (b, e, h) UoA120 × ALP731: 16 °C (n = 8), 20 °C (n = 8), 25 °C (n = 31), 30 °C (n = 8), 33 °C (n = 8). (c, f, i) UoA99 × UoA123 (ade6-M216 × ade6-3049, grey line): 16 °C (n = 18), 20 °C (n = 12), 25 °C (n = 12), 30 °C (n = 17), 33 °C (n = 17); ALP733 × ALP731 (ade6-3083 × ade6-469, black line): 11 °C (n = 11), 16 °C (n = 12), 20 °C (n = 14), 25 °C (n = 20), 30 °C (n = 12), 33 °C (n = 11); ALP1541 × ALP731 (ade6-M375 × ade6-469, green line) 16 °C (n = 12), 20 °C (n = 12), 25 °C (n = 16), 30 °C (n = 12), 33 °C (n = 11). In (c) values for the green line are to be read from green secondary y-axis. n indicates the number of independent crosses. For details of data see Table S4
The meiotic recombination assay
Our meiotic recombination assay (Osman et al. 2003; Lorenz et al. 2014; Brown et al. 2019) consists of intragenic markers (point mutations in the ade6 gene) and the intergenic markers ura4+-aim2 and his3+-aim, which are introduced to flank the ade6 gene (Fig. 2). With this assay, various recombination outcomes can be monitored simultaneously: (I) intragenic recombination (gene conversion) events producing Ade+ recombinants, (II) COs between the flanking markers and (III) the ratio of COs vs. NCOs among ade6+ gene conversions (Fig. 2a). Gene conversion and overall CO rates are determined by DSB frequencies and the choice of repair template (homologous chromosome vs. sister chromatid). After strand exchange between homologous chromosomes and recombination intermediate processing, an ade6+ gene conversion event (non-reciprocal exchange of hereditary information) can be produced, which may be accompanied by either a CO or a NCO.
Meiotic intragenic recombination levels vary greatly within the fertile range
To assess whether temperature alters meiotic recombination outcome, assays were performed at temperatures within the fertile range (Figs 2 and 3). We tested five different combinations of ade6 heteroalleles: two large intragenic intervals containing an M26-type hotspot allele (ade6-3083 × ade6-469, 1320 bp and ade6-M216 × ade6-3049, 1168 bp), one large intragenic interval containing non-hotspot alleles only (ade6-M375 × ade6-469, 1335 bp) and two small intragenic intervals containing an M26-type hotspot allele (ade6-M216 × ade6-3083, 85 bp and ade6-3049 × ade6-469, 254 bp) (Fig. 2b) (Lorenz et al. 2010, 2012; Brown et al. 2019). The ade6-M26 hotspot and its variants, ade6-3049, ade6-3083 and ade6-3074, are a well-studied type of meiotic recombination hotspot (Steiner and Smith 2005; Wahls and Davidson 2012). They contain the DNA sequence 5’-ATGACGT-3’ which generates a binding site for the transcription factor Atf1-Pcr1 (reviewed in Wahls and Davidson 2012).
The frequencies of gene conversion at both M26-type hotspots and the ade6-M375 non-hotspot are considerably lower at colder temperatures (11 °C, 16 °C and 20 °C), and tend to plateau between 25 and 33 °C (Fig. 3a–c). One of the large intervals (ade6–3083 × ade6-469) displayed a distinct peak at 30 °C (p = 2.67 × 10−11 25 °C vs. 30 °C, p = 2.6 × 10−5 30 °C vs. 33 °C; two-tailed Mann-Whitney U test) (Fig.3c). Intriguingly, the fold-change in intragenic recombination frequency between 16 °C (lowest temperature tested in all intervals) and the temperature producing the highest intragenic recombination frequency is substantially lower in the cross with the ade6-M375 non-hotspot (2.7-fold) than in the crosses containing a hotspot allele (5- to 11-fold) (Table S4). This also holds true if gene conversion frequency is compared between 16 and 25 °C (the mating temperature generally used for this type of experiment): 2.4-fold change in ade6-M375 × ade6-469 vs. a 4.3- to 6.6-fold change in the crosses containing a hotspot allele (Table S4). The very short ade6-M216 × ade6-3083 intragenic interval (85 bp) shows a stronger fold-change over temperature (6.6-fold at 16 °C vs. 25 °C), than the longer intervals containing a hotspot allele (254–1320 bp; 4.3- to 4.8-fold at 16 °C vs. 25 °C) (Table S4). This suggests, (I) that, as a general trend, lower temperatures reduce the frequency of intragenic recombination regardless of physical distance between ade6 mutations, (II) that gene conversion at the ade6-M375 non-hotspots is less sensitive to temperature changes than at hotspots and (III) that intragenic recombination at very short intervals within ade6 is singularly susceptible to temperature changes.
Meiotic CO frequency varies moderately within the fertile range
Given that major changes in gene conversion, levels are observed across temperatures, we were surprised to find that both the overall CO levels and the CO frequencies among intragenic Ade + events were less sensitive to temperature changes. The frequency of COs between ura4 + -aim2 and his3 + -aim are not substantially altered as temperature changes (Fig. 3d–f). In all intervals tested, CO frequency in the total population is only significantly lower at the temperatures of 11 and 16 °C, but then plateaus at 20 °C and higher (Fig. 3d–f, Table S5; Tukey’s Honest Significant Differences). CO frequency among ade6+ gene conversion events was even more stable with temperature changes (Fig. 3g–i). The non-hotspot only cross ade6-M375 × ade6-469 was completely unfazed by temperature changes (p = 0.314, Kruskal-Wallis test; Table S5). The crosses at cold temperatures (11, 16 and 20 °C) in all the other intervals displayed a moderate tendency to higher CO percentages than crosses at 30 °C or 33 °C (Fig. 3g–i, Table S5; Tukey’s Honest Significant Differences). The latter observation could be explained by a mechanism like CO homeostasis (Martini et al. 2006; Kan et al. 2011).
Meiotic DSB levels do not appear to change with temperature
Following the observation that temperature modulates meiotic recombination outcome, we next sought to pinpoint which specific steps during meiotic recombination are sensitive to temperature changes. Because Sz. pombe does not have a canonical synaptonemal complex (Loidl 2006), other meiotic features must be responsible for the changes in recombination outcome in response to temperature (Morgan et al. 2017). First, we assessed whether DSB formation is perturbed using the cytological markers Rec7-GFP and Rad11-GFP. Rec7 (Rec114 in S. cerevisiae), one of the co-factors essential for Spo11 recruitment and function (Molnar et al. 2001; Miyoshi et al. 2013), can be detected on meiotic chromatin and is considered a marker for DSB initiation sites (Lorenz et al. 2006). As part of RPA (replication protein A), Rad11 becomes associated with the single-stranded DNA exposed by strand resection following removal of Spo11, and is, thus, a marker for DSB formation (Parker et al. 1997). Thus, Rec7- and Rad11-focus numbers allow the indirect assessment of meiotic DSB levels. For Rec7- and Rad11-focus counts, linear elements outlined by myc-tagged Hop1 were used to identify meiotic prophase I nuclei in chromatin spreads from meiotic time-courses (Lorenz et al. 2004, 2006; Loidl and Lorenz 2009; Brown et al. 2018). To avoid potential issues of tags affecting protein functionality, we employed all tagged genes heterozygously (Lorenz et al. 2006; Brown et al. 2018); both strains (UoA825 and UoA826) behaved normally in terms of meiotic progression and sporulation efficiency during meiotic timecourses (data not shown). We chose to perform these experiments at the extreme temperatures of the fertile range (16 and 33 °C), which result in significantly different recombination frequencies at a high sporulation efficiency (Figs. 1 and 3).
Based on previous observations that recombination markers are most abundant in the thread and network stage of linear element formation (Lorenz et al. 2006), we selectively counted foci at these stages. On average ~ 16 foci per nucleus of Rec7-GFP and Rad11-GFP were observed at 16 and 33 °C (Fig. 4). The Rec7-GFP focus count was actually somewhat higher at the lower temperature (18.2 at 16 °C vs. 14.1 at 33 °C, p = 0.0017, two-tailed Mann-Whitney U test), whereas the Rad11-GFP focus numbers were indiscernible between 16 °C (15.9 foci/nucleus) and 33 °C (16.2 foci/nucleus) (Fig. 4, Table S6; p = 0.794, two-tailed Mann-Whitney U test).
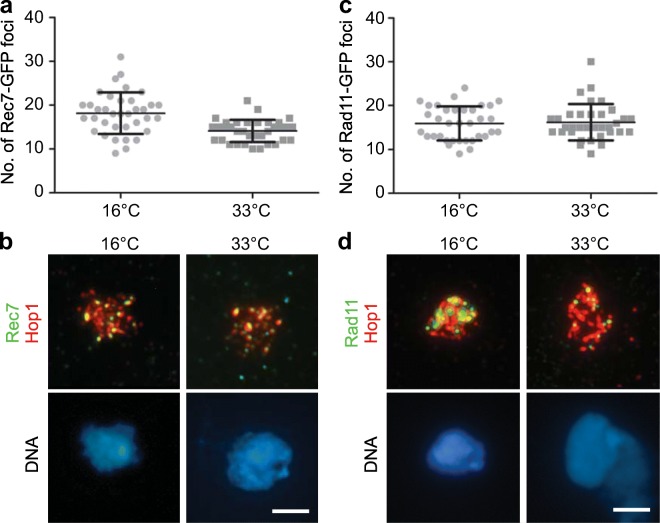
Fig. 4.
DSB formation does not seem to be affected by temperature. Focus counts of immune-detected Rec7-GFP and Rad11-GFP on Hop1-positive nuclear spreads from meiotic, azygotic timecourses at timepoints with maximum horsetail (meiotic prophase I) nucleus frequency; black horizontal lines indicate mean values, error bars represent standard deviation; for details of data see Table S6. a Number of Rec7-GFP foci per nucleus at 16 °C (25 h timepoint, n = 36) and 33 °C (5 h timepoint, n = 35) from meiotic timecourses of UoA825. b Examples of chromatin spreads evaluated in (a), Rec7-GFP in green, Hop1-13myc in red and DNA stained by Hoechst 33342 in blue; scale bar represents 5 μm. c Number of Rad11-GFP foci per nucleus at 16 °C (25 h timepoint, n = 35) and 33 °C (5 h timepoint, n = 35) from meiotic timecourses of UoA826. d Examples of chromatin spreads evaluated in (c), Rad11-GFP in green, Hop1-13myc in red and DNA stained by Hoechst 33342 in blue; scale bar represents 5 μm
These experiments suggest that overall DSB formation is possibly unaltered between 16 and 33 °C, because any of the subtle changes observed are unlikely to explain the lowered recombination frequencies at cold temperatures.
Processing of DSBs is potentially altered by temperature
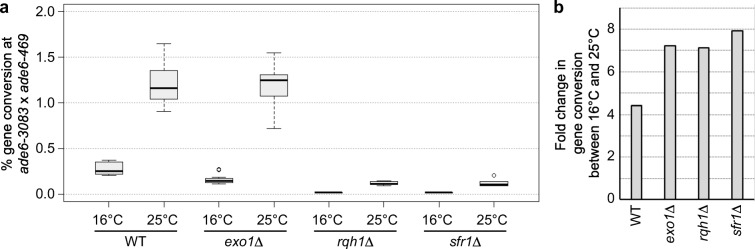
Rqh1 and Exo1 function in long-range strand resection in mitosis and meiosis in fission yeast (Langerak et al. 2011; Osman et al. 2016). Sfr1 forms a complex with Swi5 to support strand exchange, thereby promoting meiotic recombination (Ellermeier et al. 2004; Haruta et al. 2006; Lorenz et al. 2012). Less efficient DNA resection and/or reduced strand exchange with the homologous chromosomes could potentially explain why recombination levels are reduced at colder temperatures. The expectation would be that mutants defective in strand resection or strand exchange would be more sensitive to temperature changes than wild type (i.e. a synergistic effect of mutational and environmental weakening of these processes). Therefore, meiotic recombination outcome in ade6-3083 × ade6-469 crosses of rqh1Δ, exo1Δ and sfr1Δ single mutants performed at 16 °C, and 25 °C was determined. The fold difference in intragenic recombination frequency between 16 and 25 °C for wild type and each deletion was calculated to assess whether the reduction in gene conversion rate at cold temperatures is epistatic or synergistic with deleting rqh1, exo1 or sfr1 (Fig. 5). In wild-type crosses, gene conversion is 4.3-fold lower at 16 °C compared with 25 °C (p = 6.428 × 10−12, two-tailed Mann-Whitney U test). However, in rqh1Δ, exo1Δ and sfr1Δ crosses intragenic recombination levels are 7.2-fold (p = 1.402 × 10−9, two-tailed Mann-Whitney U test), 7.1-fold (p = 4.665 × 10−11, two-tailed Mann-Whitney U test) and 7.9-fold (p = 6.265 × 10−7, two-tailed Mann-Whitney U test) lower at 16 °C than at 25 °C, respectively (Fig. 5). The fold changes in overall CO frequency, and CO levels among Ade+ recombinants are largely unchanged or do not follow an obvious pattern (Table S4). Long-range strand resection and the action of strand exchange factors are potentially important for maintaining intragenic recombination frequency especially at colder temperatures, suggesting that these processes possibly are temperature-sensitive.
Fig. 5.
Cold temperature causes stronger reductions in gene conversion frequency in exo1, rqh1 or sfr1 deletions than in wild type. a Frequency of gene conversion at ade6-3083 × ade6-469 at 16 °C and 25 °C in wild type (WT), exo1Δ, rqh1Δ and sfr1Δ. ALP733 × ALP731 (WT; 16 °C n = 12, 25 °C n = 20), MCW4269 × MCW4268 (exo1Δ; 16 °C n = 11, 25 °C n = 11), ALP781 × ALP780 (rqh1Δ; 16 °C n = 12, 25 °C n = 10), ALP800 × ALP782 (sfr1Δ; 16 °C n = 11, 25 °C n = 10). n indicates the number of independent crosses. The upper, middle and lower lines of the box represent the third, second and first quartile, respectively (second quartile = median). The ‘whiskers’ represent the minimum and maximum of the range, unless they differ more than 1.5-times the interquartile distance from the median, then the 1.5-times interquartile distance around the median is indicated by the ‘whiskers’, and outliers are shown as open circles. b Fold change of data in (a). For details of data see Table S4
Discussion
Generally, meiosis happens faster at higher temperatures, this has been observed in multiple (model) organisms (Bennett 1977; De La Peña et al. 1980; Stefani and Colonna 1996; Bomblies et al. 2015). However, this gain in developmental speed comes at the expense of fidelity indicated by cellular abnormalities and reduced reproductive success at higher temperatures (Loidl 1989; Stefani and Colonna 1996; Bomblies et al. 2015). In addition to pathological changes to the chromosome axis (Loidl 1989; Higgins et al. 2012; Morgan et al. 2017), the processing of recombination intermediates has been highlighted as a temperature-sensitive mechanism. The ability of meiotic RecA-family protein isolated from mouse and lily to support D-loop formation in vitro is decreased at higher temperatures (23 °C against 33 °C for lily, and 33 °C against 37 °C for mouse) (Hotta et al. 1985). This temperature preference of meiotic recombinase protein isolated from mouse aligns nicely with the temperature of mammalian testes in the scrotum, which tends to be ~ 5 °C lower than the normal body temperature of ~ 37 °C. Indeed, artificial cryptorchidism generated by surgically transplanting testes into the abdominal cavity of rats (i.e. raising testis temperature to body temperature in an in vivo system) causes substantial drops in recombination activity (Hotta et al. 1988).
The environmental temperature regime during sexual reproduction also influences meiotic recombination outcome (Bomblies et al. 2015). Temperature effects on recombination are largely species-specific and can manifest in different ways: (I) CO frequencies follow a U-shaped distribution in Drosophila or Arabidopsis (Plough 1917; Lloyd et al. 2018), where CO recombination is highest at the more extreme temperatures within the fertile range, (II) CO frequency increases with increasing temperature in C. elegans (Rose and Baillie 1979), (III) in grasshoppers CO frequency decreases with increasing temperature (Church and Wimber 1969) and (IV) in S. cerevisiae, Hordeum vulgare (barley) and Secale cereale (rye) overall CO frequency is maintained (De La Peña et al. 1980; Higgins et al. 2012; Zhang et al. 2017). In the latter situation, CO positioning can be altered, as demonstrated for budding yeast and barley (Higgins et al. 2012; Zhang et al. 2017). In S. cerevisiae this has largely been explained by DSB hotspot activation varying with temperature (Zhang et al. 2017).
In fission yeast, changes in environmental temperature during sexual reproduction affect the duration of mating, meiosis and sporulation (Fig. 1), as well as the meiotic recombination outcome tested at several versions of a genetic interval containing multiple different ade6 heteroalleles (Fig. 3). The fertile range of Sz. pombe extends from ~ 10 to ~ 33 °C (Fig. 1). The duration of sexual reproduction lasts from 2 weeks at 11 °C to 48 h at 33 °C (Fig. 1). The CO frequency changes over temperature observed between ura4+-aim2 and his3+-aim (Fig. 3d–f) do definitely not follow a U-shaped distribution like in Drosophila or Arabidopsis (Plough 1917; Lloyd et al. 2018), but are similar to C. elegans (Rose and Baillie 1979) where CO rates tend to be lower at low temperatures (Fig. 3d–f). Interestingly, gene conversion frequency at various different ade6 heteroalleles shows strong changes with temperature within the fertile range, this is considerably more affected than COs and COs among intragenic events between ura4+-aim2 and his3+-aim (Fig. 3). Within the fertile range, overall DSB levels appear to be similar at 16 °C and 33 °C in fission yeast as estimated by immunocytochemistry of Rec7-GFP and Rad11-GFP (Fig. 4). We counted an average of 14–18 foci/nucleus for both cytological markers in Hop1-positive nuclei; Hop1-staining of the meiotic chromosome axis was used to determine that the analysed nuclei were at the appropriate stage with the highest recombination events (Lorenz et al. 2004; Brown et al. 2018). This is similar to average focus counts for Rec7-GFP and Rad51 in a previous study (Lorenz et al. 2006), and is in line with an estimated ~ 60 DSB events/nucleus/meiosis (Fowler et al. 2014) considering that single-DSB events might cluster together (Fowler et al. 2018). Previously, temperature-induced changes in physical DSB formation at an ade6 hotspot allele have been observed in a unisexual and artificially synchronized meiotic system using mutant pat1 alleles, with DSB frequency being notably lower at 25 °C than at 34 °C (Hyppa et al. 2014). These discrepancies can have several technical sources, such as higher synchrony in meiotic cultures of pat1 strains (Loidl and Lorenz 2009), and the unisexuality of the pat1 synchronization tool affecting various meiotic processes (Bähler et al. 1991; Chikashige et al. 2004; Hyppa and Smith 2010; Hyppa et al. 2014) resulting in different readouts compared with normal zygotic or azygotic meiosis. Additionally, these discrepancies can point towards genuine biological differences, i.e. that DSB frequency is indeed altered between 25 and 34 °C at particular M26-type ade6 hotspots, which might not be detectable in recombination outcome due to CO homeostasis and/or CO invariance (Hyppa and Smith 2010; Kan et al. 2011), whereas the more drastic reductions in gene conversion frequencies seen at 16 °C are caused by inefficiencies in DSB processing (Fig. 5). These two observations are also not mutually exclusive and could well overlap over the full fertile range. Further in-depth experimentation will be required to determine the exact contribution of these mechanisms to the thermotolerance of Sz. pombe meiosis. Considering that M26-type hotspots require the transcription factor Atf1-Pcr1 for full activation (Kon et al. 1997), any temperature-induced changes could also be caused by alterations in Atf1-Pcr1 recruitment to the hotspot sequence; this remains to be tested experimentally. In contrast to S. cerevisiae where ~ 80% of DSBs change location at different temperatures (14, 30 and 37 °C) (Zhang et al. 2017), only 17 out of 288 DSB hotspots behaved differentially between 25 and 34 °C in Sz. pombe (Hyppa et al. 2014); it is, thus, less likely that activation of different hotspots is a major confounding factor in changing of recombination frequency at a given site in Sz. pombe. Considering that overall CO frequency between ura4+-aim2 and his3+-aim is only moderately affected by temperature in a given interval, whereas gene conversion rates at ade6 change massively, a switch from interhomologue to intersister recombination is also an unlikely factor, since it would affect intergenic COs and intragenic recombination to a similar extent. This discrepancy between gene conversion and CO frequency changes in relation to environmental temperature needs to be resolved. Our observation that strand resection, strand exchange and/or branch migration seem to be compromised at colder temperatures (Fig. 5), indicates that inefficiencies in these processes impair the conversion of point mutations in ade6 alleles to wild-type ade6+. Nevertheless, mature recombination intermediates (D-loops, Holliday Junctions) can be formed between homologous chromosomes at these sites even at low temperatures, thus maintaining CO frequency. Furthermore, recombination monitored at the ade6-M375 non-hotspot allele is less sensitive to temperature changes than that involving an M26-type ade6 hotspot (Fig. 3, Table S4). This indifference to temperature could be a manifestation of CO invariance, which has been suggested as an explanation for a stronger drive towards interhomologue recombination in regions of low meiotic recombination frequency (Hyppa and Smith 2010).
One of the most obvious traits of fission yeast meiosis is the lack of a synaptonemal complex (Loidl 2006). Because extreme temperatures affect synaptonemal complex structure (Loidl 1989; Higgins et al. 2012; Bilgir et al. 2013), and mutants of components of the synaptonemal complex or the meiotic chromosome axis show differential phenotypes at different temperatures (Börner et al. 2004; Penedos et al. 2015), it has been suggested that the synaptonemal complex is a salient factor in determining the meiotic thermotolerance of a species (Morgan et al. 2017). In Sz. pombe, though, meiotic thermotolerance seems to rely on biochemical activities supporting DSB formation (Hyppa et al. 2014), recombination intermediate formation (DNA strand exchange) and/or stabilization (Fig. 5). Although, Sz. pombe lacks a fully-fledged synaptonemal complex, components of its linear elements are homologous to lateral element proteins of the synaptonemal complex (Lorenz et al. 2004; Loidl 2006). It is rather difficult to test whether aspects of meiotic thermotolerance are conferred by the linear elements, because deletion of linear element genes strongly reduces meiotic recombination or, in most cases, abolishes it completely (Ellermeier and Smith 2005; Davis et al. 2008; Latypov et al. 2010; Estreicher et al. 2012). However, rec10–144, a hypomorphic mutant of the main linear element component rec10, shows some temperature sensitivity in intragenic recombination (Pryce et al. 2005). This suggests that in addition to DSB formation and processing, linear elements might also play a role in meiotic thermotolerance.
Despite more than 100 years of research (Plough 1917), we still only have a basic understanding of how environmental parameters influence meiotic recombination outcomes in different species. Environmental temperature grossly affects the speed of sexual reproduction (Bennett 1977) and recombination levels in several species (Plough 1917; Rose and Baillie 1979; Bomblies et al. 2015; Lloyd et al. 2018), likely by changing the frequency of DSB formation at a given site (Hyppa et al. 2014), the positioning of the initial DSB (Higgins et al. 2012; Zhang et al. 2017) and/or dynamics of DSB repair (Modliszewski et al. 2018; this study). Here, we define the fertile range over temperature in the fission yeast Schizosaccharomyces pombe, and determine that this stipulates the pace of sexual reproduction and the level of recombination, gene conversion events in particular. At least in part, the latter seems to be driven by temperature-sensitive steps of recombination intermediate processing downstream of DSB formation, such as DNA strand exchange and/or branch migration. Our study highlights the importance of the interplay between intrinsic and environmental factors in shaping the genetic diversity of a given population, and the need for further experimentation to elucidate the cellular mechanisms underpinning meiotic thermotolerance.
Electronic supplementary material
(XLSX 39 kb)
Acknowledgements
We are grateful to Miguel G. Ferreira, Jürg Kohli, Josef Loidl, Fekret Osman, Gerald R. Smith, Walter W. Steiner and Matthew Whitby for providing strains. We thank Josef Loidl for critically reading an earlier version of this manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council UK (BBSRC) [grant number BB/F016964/1] and the University of Aberdeen (College of Life Sciences and Medicine Start-up grant to AL).
Abbreviations
- aim
artificially introduced marker
- DSB(s)
DNA double-strand break(s)
- DNA
deoxyribonucleic acid
- CO(s)
crossover(s)
- GC-CO
gene conversion associated with a crossover
- GC-NCO
gene conversion associated with a non-crossover
- GFP
green fluorescent protein
- ME
malt extract
- NCO(s)
non-crossover(s)
- YE
yeast extract
- YNG
yeast nitrogen base glutamate
Author contributions
SDB: conception and design, unpublished essential reagents (yeast strains, plasmids), acquisition of data, analysis and interpretation of data, and revising the manuscript; CA: acquisition of data and revising the manuscript; AL: conception and design, unpublished essential reagents (yeast strains, plasmids), acquisition of data, analysis and interpretation of data and drafting and revising the manuscript.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bähler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr Genet. 1991;19:445–451. doi: 10.1007/bf00312735. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD. The time and duration of meiosis. Philos Trans R Soc Lond B Biol Sci. 1977;277:201–226. doi: 10.1098/rstb.1977.0012. [DOI] [PubMed] [Google Scholar]
- Bilgir C, Dombecki CR, Chen PF, Villeneuve AM, Nabeshima K. Assembly of the synaptonemal complex is a highly temperature-sensitive process that is supported by PGL-1 during Caenorhabditis elegans meiosis. G3 (Bethesda) 2013;3:585–595. doi: 10.1534/g3.112.005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Higgins JD, Yant L. Meiosis evolves: adaptation to external and internal environments. New Phytol. 2015;208:306–323. doi: 10.1111/nph.13499. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/S0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Brown SD, Jarosinska OD, Lorenz A. Genetic interactions between the chromosome axis-associated protein Hop1 and homologous recombination determinants in Schizosaccharomyces pombe. Curr Genet. 2018;64:1089–1104. doi: 10.1007/s00294-018-0827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Mpaulo SJ, Asogwa MN, Jézéquel M, Whitby MC, Lorenz A. DNA sequence differences are determinants of meiotic recombination outcome. Sci Rep. 2019;9:16446. doi: 10.1038/s41598-019-52907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2004;9:671–684. doi: 10.1111/j.1356-9597.2004.00760.x. [DOI] [PubMed] [Google Scholar]
- Church K, Wimber DE. Meiosis in the grasshopper: chiasma frequency after elevated temperature and X-rays. Can J Genet Cytol. 1969;11:209–216. doi: 10.1139/g69-025. [DOI] [PubMed] [Google Scholar]
- Davis L, Rozalén AE, Moreno S, Smith GR, Martín-Castellanos C. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Peña A, Yllera P, Puertas MJ. Effect of temperature on chromosome behaviour of in vitro cultivated anthers of rye. Caryologia. 1980;33:411–418. doi: 10.1080/00087114.1980.10796854. [DOI] [Google Scholar]
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- Ellermeier C, Schmidt H, Smith GR. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estreicher A, Lorenz A, Loidl J. Mug20, a novel protein associated with linear elements in fission yeast meiosis. Curr Genet. 2012;58:119–127. doi: 10.1007/s00294-012-0369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-Parra I, Zhurinsky J, Bernal M, Gallardo P, Daga RR. A Lallzyme MMX-based rapid method for fission yeast protoplast preparation. Yeast. 2014;31:61–66. doi: 10.1002/yea.2994. [DOI] [PubMed] [Google Scholar]
- Fowler KR, Hyppa RW, Cromie GA, Smith GR. Physical basis for long-distance communication along meiotic chromosomes. Proc Natl Acad Sci U S A. 2018;115:E9333–E9342. doi: 10.1073/pnas.1801920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Cohen PE. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev Genet. 2016;50:175–210. doi: 10.1146/annurev-genet-120215-035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FC. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell. 2012;24:4096–4109. doi: 10.1105/tpc.112.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Fujisawa M, Tabata S, Stern H, Yoshida S. The effect of temperature on recombination activity in testes of rodents. Exp Cell Res. 1988;178:163–168. doi: 10.1016/0014-4827(88)90387-4. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Tabata S, Bouchard RA, Piñon R, Stern H. General recombination mechanisms in extracts of meiotic cells. Chromosoma. 1985;93:140–151. doi: 10.1007/BF00293161. [DOI] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Fowler KR, Cipak L, Gregan J, Smith GR. DNA intermediates of meiotic recombination in synchronous S. pombe at optimal temperature. Nucleic Acids Res. 2014;42:359–369. doi: 10.1093/nar/gkt861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC. The natural diversity and ecology of fission yeast. Yeast. 2018;35:253–260. doi: 10.1002/yea.3293. [DOI] [PubMed] [Google Scholar]
- Kan F, Davidson MK, Wahls WP. Meiotic recombination protein Rec12: functional conservation, crossover homeostasis and early crossover/non-crossover decision. Nucleic Acids Res. 2011;39:1460–1472. doi: 10.1093/nar/gkq993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol Cell Biol. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife. 2015;4:e05835. doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, Franklin C, Bomblies K. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics. 2018;208:1409–1420. doi: 10.1534/genetics.117.300588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- Loidl J. Conservation and variability of meiosis across the eukaryotes. Annu Rev Genet. 2016;50:293–316. doi: 10.1146/annurev-genet-120215-035100. [DOI] [PubMed] [Google Scholar]
- Loidl J. Effects of elevated temperature on meiotic chromosome synapsis in Allium ursinum. Chromosoma. 1989;97:449–458. doi: 10.1007/BF00295029. [DOI] [Google Scholar]
- Loidl J, Lorenz A. Analysis of Schizosaccharomyces pombe meiosis by nuclear spreading. Methods Mol Biol. 2009;558:15–36. doi: 10.1007/978-1-60761-103-5_2. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Mehats A, Osman F, Whitby MC. Rad51/Dmc1 paralogs and mediators oppose DNA helicases to limit hybrid DNA formation and promote crossovers during meiotic recombination. Nucleic Acids Res. 2014;42:13723–13735. doi: 10.1093/nar/gku1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F, McFarlane R, Loidl J. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci. 2004;117:3343–3351. doi: 10.1242/jcs.01203. [DOI] [PubMed] [Google Scholar]
- Lorenz A, West SC, Whitby MC. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 2010;38:1866–1873. doi: 10.1093/nar/gkp1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BC. Genetic recombination in Coprinus. IV a kinetic study of the temperature effect on recombination frequency. Genetics. 1974;78:661–677. doi: 10.1093/genetics/78.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Ohta K. Spatiotemporal regulation of meiotic recombination by Liaisonin. Bioarchitecture. 2013;3:20–24. doi: 10.4161/bioa.23966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modliszewski JL, Wang H, Albright AR, Lewis SM, Bennett AR, Huang J, Ma H, Wang Y, Copenhaver GP. Elevated temperature increases meiotic crossover frequency via the interfering (type I) pathway in Arabidopsis thaliana. PLoS Genet. 2018;14:e1007384. doi: 10.1371/journal.pgen.1007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci. 2003;116:1719–1731. doi: 10.1242/jcs.00387. [DOI] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CH, Zhang H, Bomblies K. Are the effects of elevated temperature on meiotic recombination and thermotolerance linked via the axis and synaptonemal complex? Philos Trans R Soc Lond B Biol Sc. 2017;372:20160470. doi: 10.1098/rstb.2016.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LW, Eden U, Egel-Mitani M, Egel R. Asynaptic meiosis in fission yeast? Hereditas. 1978;89:189–199. doi: 10.1111/j.1601-5223.1978.tb01275.x. [DOI] [Google Scholar]
- Osman F, Ahn JS, Lorenz A, Whitby MC. The RecQ DNA helicase Rqh1 constrains exonuclease 1-dependent recombination at stalled replication forks. Sci Rep. 2016;6:22837. doi: 10.1038/srep22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/S1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Parker AE, Clyne RK, Carr AM, Kelly TJ. The Schizosaccharomyces pombe rad11+ gene encodes the large subunit of replication protein a. Mol Cell Biol. 1997;17:2381–2390. doi: 10.1128/MCB.17.5.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedos A, Johnson AL, Strong E, Goldman AS, Carballo JA, Cha RS. Essential and checkpoint functions of budding yeast ATM and ATR during meiotic prophase are facilitated by differential phosphorylation of a meiotic adaptor protein, Hop1. PLoS One. 2015;10:e0134297. doi: 10.1371/journal.pone.0134297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips Dylan, Jenkins Glyn, Macaulay Malcolm, Nibau Candida, Wnetrzak Joanna, Fallding Derek, Colas Isabelle, Oakey Helena, Waugh Robbie, Ramsay Luke. The effect of temperature on the male and female recombination landscape of barley. New Phytologist. 2015;208(2):421–429. doi: 10.1111/nph.13548. [DOI] [PubMed] [Google Scholar]
- Plough HH. The effect of temperature on linkage in the second chromosome of Drosophila. Proc Natl Acad Sci U S A. 1917;3:553–555. doi: 10.1073/pnas.3.9.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce DW, Lorenz A, Smirnova JB, Loidl J, McFarlane R. Differential activation of M26-containing meiotic recombination hot spots in Schizosaccharomyces pombe. Genetics. 2005;170:95–106. doi: 10.1534/genetics.104.036301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AM, Baillie DL. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics. 1979;92:409–418. doi: 10.1093/genetics/92.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- Stefani A, Colonna N. The influence of temperature on meiosis and microspores development in Dasypyrum villosum (L.) P. Candargy. Cytologia (Tokyo) 1996;61:277–283. doi: 10.1508/cytologia.61.277. [DOI] [Google Scholar]
- Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock AJ, Henderson IR. Hotspots for initiation of meiotic recombination. Front Genet. 2018;9:521. doi: 10.3389/fgene.2018.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Davidson MK. New paradigms for conserved, multifactorial, cis-acting regulation of meiotic recombination. Nucleic Acids Res. 2012;40:9983–9989. doi: 10.1093/nar/gks761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wu X-C, Zheng D-Q, Petes TD. Effects of temperature on the meiotic recombination landscape of the yeast Saccharomyces cerevisiae. MBio. 2017;8:e02099–e02017. doi: 10.1128/mBio.02099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 39 kb)