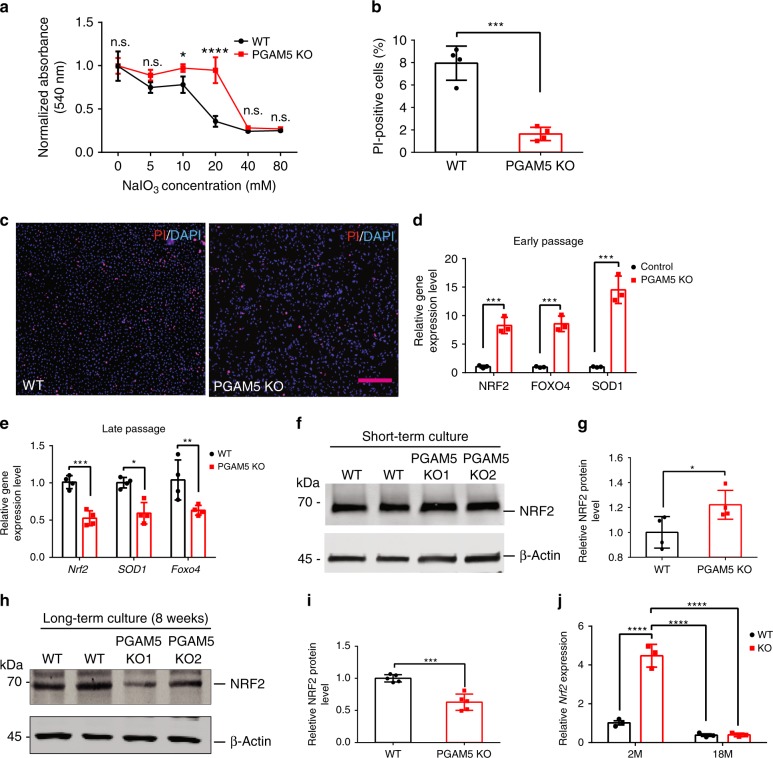
Fig. 8. Age-dependent regulation of anti-oxidative genes by PGAM5.
a Cell survival by neutral red assay in WT and PGAM5−/− ARPE-19 cells after overnight NaIO3 treatment using indicated concentrations. n = 4, *p < 0.05, ****p < 0.0001, two-way ANOVA Tukey’s multiple comparisons test. Error bars, mean ± s.d. b Quantification of PI staining showing cell death in WT and PGAM5−/− ARPE-19 cells after overnight NaIO3 (10 mM) treatment using Nikon Elements software and ImageJ. n = 4, ***p = 0.0002, two-tailed unpaired t-tests, error bars, mean ± s.d. c Representative PI staining in b. Scale bar = 250 µm. n = 4. d Upregulation of anti-oxidative genes (NRF2, FOXO4 and SOD1) in PGAM5−/− ARPE-19 cells compared to control cells after short-term (1 week) culture. Data were normalized to GAPDH. n = 3, ***p = 0.001(Nrf2), p = 0.0007(Foxo4), p = 0.0006, two-tailed unpaired t-tests, error bars, mean ± s.d. e Downregulation of anti-oxidative genes (NRF2, FOXO4 and SOD1) in PGAM5−/− ARPE-19 cells compared to control cells after long-term (8 weeks) culture. Data were normalized to GAPDH. n = 4, ***p = 0.0003; **p = 0.0022; *p = 0.0258, two-tailed unpaired t-tests, error bars, mean ± s.d. f Upregulation of NRF2 protein in PGAM5−/− ARPE-19 cells compared to control cells after short-term (1 week) culture. Data were normalized to β-actin. n = 4. g Quantification of data in f. n = 4, *p = 0.042, two-tailed unpaired t-tests, error bars, mean ± s.d. h Downregulation of NRF2 protein in PGAM5−/− ARPE-19 cells compared to control cells after long-term (8 weeks) culture. Data were normalized to β-actin. n = 5. i Quantification of data in h. n = 5, ***p = 0.0003, two-tailed unpaired t-tests, error bars, mean ± s.d. j Nrf2 expression as measured by qRT-PCR in the RPE/choroid of WT and Pgam5−/− mice at 2 and 18-months old. Data were normalized to Gapdh. n = 3, ****p < 0.0001, two-way ANOVA Tukey’s multiple comparisons test. Error bars, mean ± s.d. For assays in the figure, n represents the number of biologically independent experiments unless otherwise specified. Images were captured under same settings, and representative images were shown. Source data are available as a Source Data file.