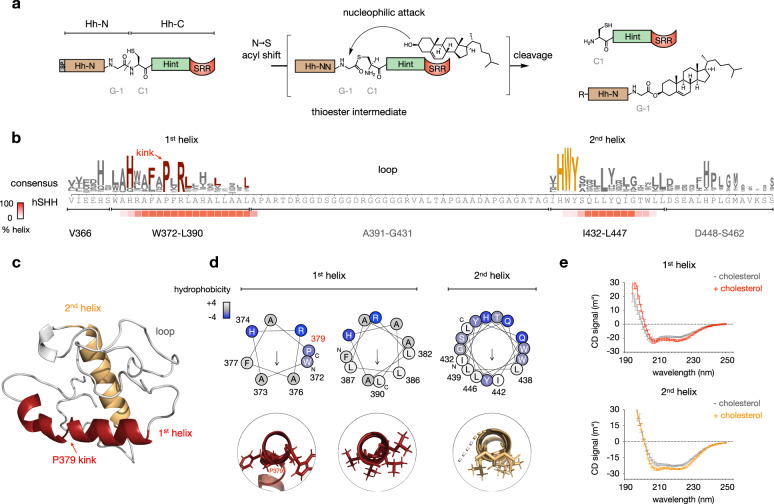
Fig. 1. The SRR is a helix-loop-helix motif.
a The Hint domain within full-length Hedgehog (Hh) proteins catalyzes formation of a thioester intermediate in the peptide backbone. The SRR facilitates cholesterol attack on the thioester, cleaving the Hh protein and ligating cholesterol to the last residue of the N-terminus. In a separate process, palmitate is attached to the first residue of the N-terminus after cleavage of the signal peptide. Conventional Hint domain numbering is shown; R = palmitoyl. b Sequence logo for PROMALS3D alignment (ref. 24) of the SRR of 700 manually annotated Hedgehog protein sequences above the corresponding sequence of hSHH (SRR, residues 363–462). Heatmap below shows the percent helical character predicted by the JNet4 algorithm (ref. 25). c Model of the SRR of hSHH (residues 363–462), created using ab initio Rosetta prediction (ref. 26). d Helical wheel diagrams of the 1st and 2nd SRR helices from HELIQUEST (ref. 26). The 1st helix is twisted at P379; axial views of each segment are shown. Residues are colored according to the Kyte Doolittle hydrophobicity scale (white = hydrophobic, blue = hydrophilic). e Circular dichroism (CD) spectra of peptides encompassing the 1st helix (residues 368–391) and 2nd helix (residues 431–449) in liposomes.