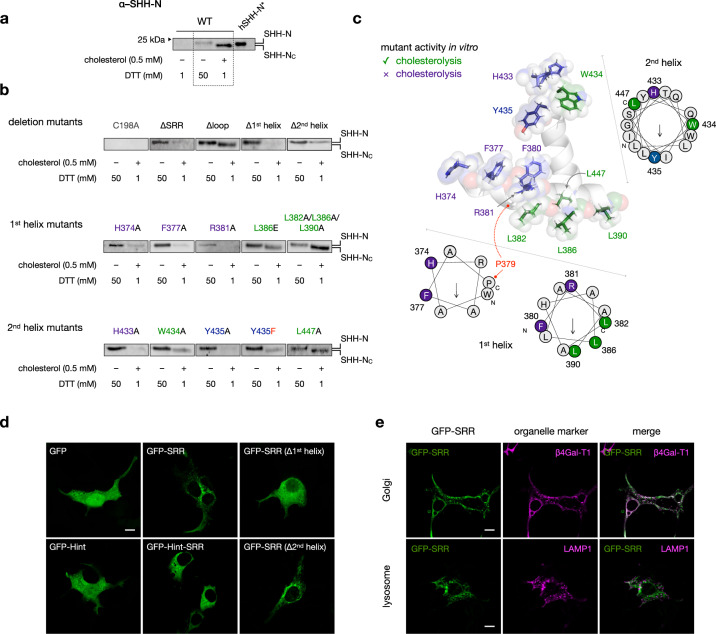
Fig. 3. Specific SRR residues control biochemical cholesterolysis and cellular localization.
a Wild-type hSHH-FL isolated from HEK293T cells shows no cleavage in 1 mM DTT, non-cholesteroylative cleavage with 50 mM DTT, and cholesteroylative cleavage with 1 mM DTT + 0.5 mM cholesterol. Cholesteroylated hSHH-N (hSHH-NC) shows a characteristic migration shift relative to non-cholesteroylated hSHH-N, as demonstrated by protein generated from a construct expressing hSHH-N only (hSHH-N*, residues 1-197). b Cleavage and/or cholesterolysis hSHH mutants by 50 mM DTT or 1 mM DTT + 0.5 mM cholesterol. For additional blots, see Supplementary Fig. 5. c SRR model showing residues required for cholesterol modification both in cells and in vitro (violet), in cells but not in vitro (green), and Y435 (blue), which is required both in cells and in vitro but can be functionally replaced by phenylalanine. Helical wheels show the facial positions of functional residues; the 1st helix is divided into coaxial segments before and after the P379 kink. d Confocal microscopy images of HEK293T cells expressing EGFP-SRR fusion proteins. e Images of EGFP-SRR coexpressed with mCherry fused to the Golgi-targeting sequence from β4-galactosyltransferase-1 (β4Gal-T1) or the lysosome-targeting sequence from LAMP1. Scale bar = 10 µM.