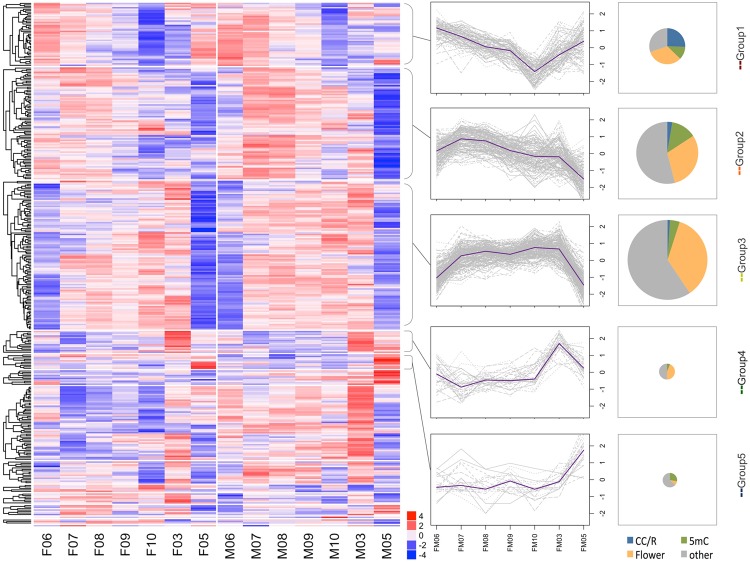
Figure 4.
Groups of chromatin modification genes show strong temporal patterns (heat map). Transcript abundance data for all expressed poplar homologs of genes in the functional category ‘covalent chromatin modification’ (GO:0016569, AMIGO db, Arabidopsis) are shown. Data were subjected to hierarchical clustering, subdivided based on major clusters, and also plotted as line graphs to visualize temporal change. Additional functional information was retrieved from recent publications and is shown as pie charts. Group1 includes DNA methylation and cell-cycle related compounds (CMT3, DDM1, KYP), histone methyltransferases (JMJ12/REF6, JMJ13, regulation of flowering); Group 2: RdDM (AGO4, DRD1, SHH1), FLD, trxG (ATX1, floral organ, flowering); Group 3: histone methyltransferases (JMJ16, JMJ20, ASHH2, flowering, organ development); Group 4: ubiquitination (UBC2, ENY), histone deacetylase (HDA19, flowering, floral patterning). Some chromatin modification genes show male bias (PRMT3,5, and 6). Interestingly very little chromatin modification activity is evident at the final time-point (mature buds, just prior to anthesis). Line plots show median (purple line) and individual (grey) expression patterns per group. The size of the pie charts reflects the number of genes in each respective group, additional biological functions curated from recent literature are color coded. CC/R: cell cycle or DNA repair, 5mC: DNA methylation at cytosine, Flower: role in the regulation of flowering, flowering time or flower organ development. Sample labels: F/M represents female or male; numbers correspond to months. E.g. F06 corresponds to female buds harvested in June.