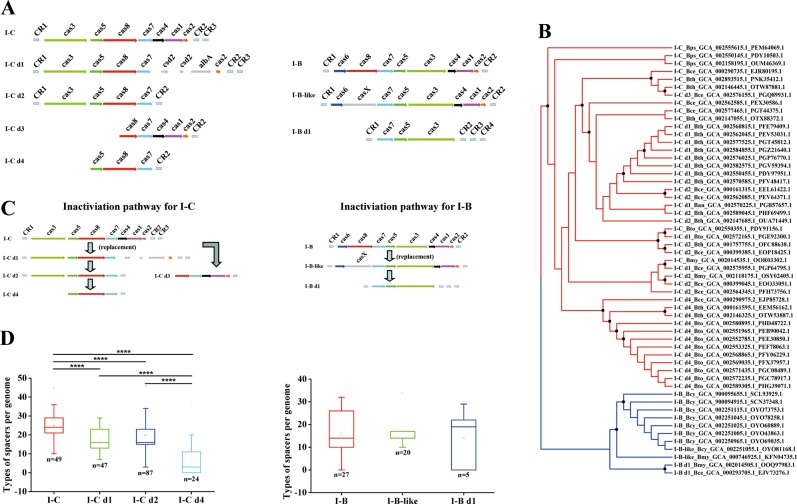
Fig. 2. Genetic inactivation of CRISPR-Cas system commonly occurs in B. cereus group.
a Comparison of the genetic organization of variants of type I CRISPR-Cas systems. All the sequences were retrieved from NCBI and their accession numbers are listed in Table S1. Only representative strains are shown. b The maximum-likelihood (ML) phylogenetic tree based on the sequence of Cas7 protein was constructed by FastTree. The strains with intact or variants of subtypes I-C systems are shown in red branches, while the intact or variants of subtypes I-B systems are shown in blue branches. Nodes supported with a bootstrap values ≥50% are indicated with a black dot. Ecotypes names of B. cereus group are abbreviated as follows: Ban for B. anthracis, Bce for B. cereus, Bcy for B. cytotoxicus, Bmy for B. mycoides, Bps for B. pseudomycoides, Bth for B. thuringiensis, and Bto for B. toyonensis. The numbers, such as GCA_002560815.1 and PFE79409.1 are the GenBank assembly accessions of genomes and GenBank accessions of Cas7 proteins, respectively. c Proposed inactivation pathways of type I CRISPR-Cas system in B. cereus group. The predicted inactivation pathways for subtypes I-C or subtypes I-B variants are shown. d Differences of spacer types among strains with variants of CRISPR-Cas system in the B. cereus group. The spacers of variant I-C d3 were excluded because only one strain contains this variant. All the spacers are listed in Dataset 1.